8YD8
 
 | |
8YD7
 
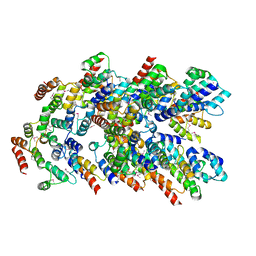 | | Structure of FADD/Caspase-8/cFLIP death effector domain assembly | | Descriptor: | CASP8 and FADD-like apoptosis regulator subunit p12, Caspase-8, FAS-associated death domain protein, ... | | Authors: | Lin, S.-C, Yang, C.-Y. | | Deposit date: | 2024-02-19 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Deciphering DED assembly mechanisms in FADD-procaspase-8-cFLIP complexes regulating apoptosis.
Nat Commun, 15, 2024
|
|
8YBX
 
 | |
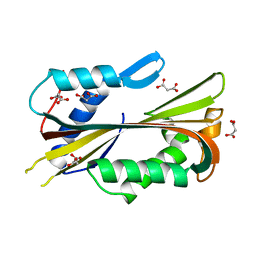
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
1VCY
 
 | | VVA2 isoform | | Descriptor: | MALONATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-03-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1VGF
 
 | | volvatoxin A2 (diamond crystal form) | | Descriptor: | ACETATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-04-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
8YM6
 
 | |
8YM4
 
 | |
8YM5
 
 | |
8YNI
 
 | |
8YNK
 
 | |
8YNL
 
 | |
8YNM
 
 | |
8YNN
 
 | |
3DKB
 
 | | Crystal Structure of A20, 2.5 angstrom | | Descriptor: | Tumor necrosis factor, alpha-induced protein 3 | | Authors: | Lin, S.-C, Chung, J.Y, Lo, Y.-C, Wu, H. | | Deposit date: | 2008-06-24 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20
J.Mol.Biol., 376, 2008
|
|
1PP0
 
 | | volvatoxin A2 in monoclinic crystal | | Descriptor: | ACETIC ACID, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1PP6
 
 | |
2QRA
 
 | | Crystal structure of XIAP BIR1 domain (P21 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ETHANOL, ZINC ION | | Authors: | Lin, S.-C. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the BIR1 domain of XIAP in two crystal forms
J.Mol.Biol., 372, 2007
|
|
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCT
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1V2G
 
 | | The L109P mutant of E. coli Thioesterase I/Protease I/Lysophospholipase L1 (TAP) in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, IMIDAZOLE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2003-10-15 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1U8U
 
 | | E. coli Thioesterase I/Protease I/Lysophospholiase L1 in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, GLYCEROL, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2004-08-07 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1JRL
 
 | | Crystal structure of E. coli Lysophospholiase L1/Acyl-CoA Thioesterase I/Protease I L109P mutant | | Descriptor: | Acyl-CoA Thioesterase I, IMIDAZOLE, SULFATE ION | | Authors: | Lo, Y.-C, Lin, S.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2001-08-14 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
