2O6H
 
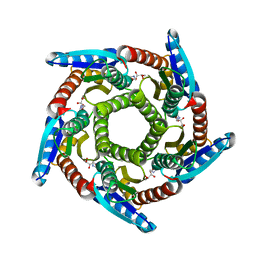 | | Lumazine synthase RibH1 from Brucella melitensis (Gene BMEI1187, Swiss-Prot entry Q8YGH2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2OBX
 
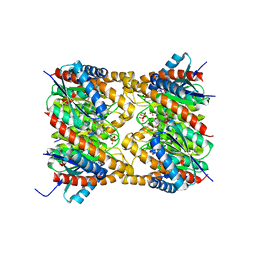 | | Lumazine synthase RibH2 from Mesorhizobium loti (Gene mll7281, Swiss-Prot entry Q986N2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and kinetic properties of lumazine synthase isoenzymes in the order rhizobiales
J.Mol.Biol., 373, 2007
|
|
2MGX
 
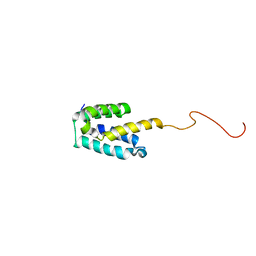 | | NMR structure of SRA1p C-terminal domain | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Bilinovich, S.M, Davis, C.M, Morris, D.L, Ray, L.A, Prokop, J.W, Buchan, G.J, Leeper, T.C. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domain of SRA1p Has a Fold More Similar to PRP18 than to an RRM and Does Not Directly Bind to the SRA1 RNA STR7 Region.
J.Mol.Biol., 426, 2014
|
|
2MZC
 
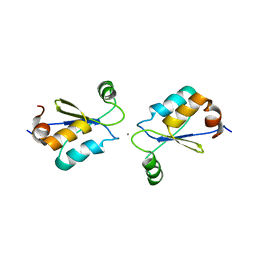 | | Metal Binding of Glutaredoxins | | Descriptor: | Glutaredoxin, SILVER ION | | Authors: | Bilinovich, S.M, Caporoso, J.A, Taraboletti, A, Duangjumpa, N, Panzner, M.J, Prokop, J.W, Shriver, L.P, Leeper, T.C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal Binding of Glutaredoxins
To be Published
|
|
4D6X
 
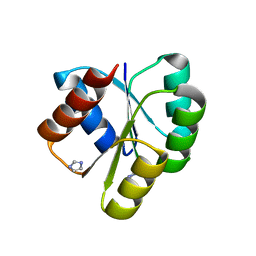 | | Crystal structure of the receiver domain of NtrX from Brucella abortus | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, IMIDAZOLE | | Authors: | Klinke, S, Fernandez, I, Carrica, M.C, Otero, L.H, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
2CXJ
 
 | |
2F59
 
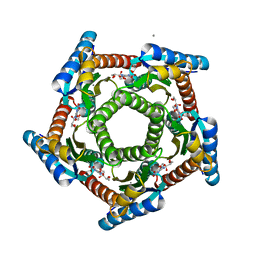 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2FUI
 
 | |
2FUU
 
 | |
5OMS
 
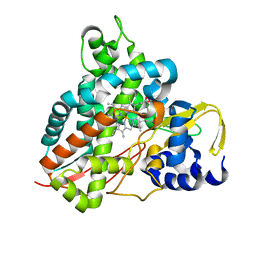 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with guaethol. | | Descriptor: | 2-ethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OGX
 
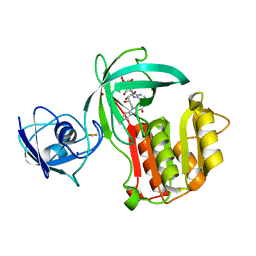 | | Crystal structure of Amycolatopsis cytochrome P450 reductase GcoB. | | Descriptor: | BROMIDE ION, Cytochrome P450 reductase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-07-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMU
 
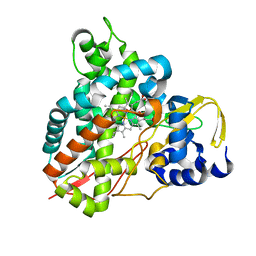 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
5OMR
 
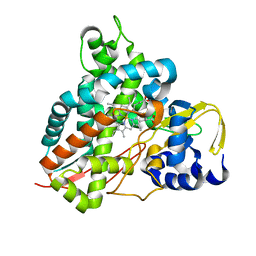 | | Crystal structure of Amycolatopsis cytochrome P450 GcoA in complex with vanillin. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, GcoA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Johnson, C.W, Neidle, E.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-08-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Nat Commun, 9, 2018
|
|
2JMI
 
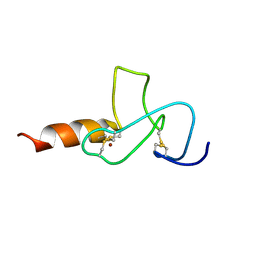 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2JMJ
 
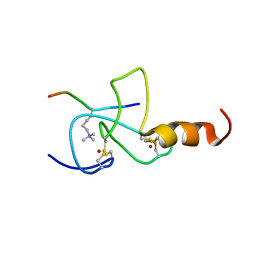 | | NMR solution structure of the PHD domain from the yeast YNG1 protein in complex with H3(1-9)K4me3 peptide | | Descriptor: | Histone H3, Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2U2A
 
 | |
2I0F
 
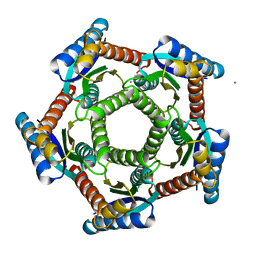 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2JYP
 
 | |
5A3I
 
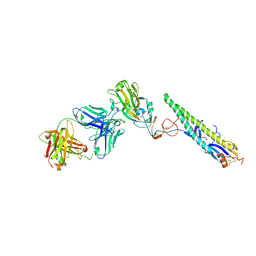 | | Crystal Structure of a Complex formed between FLD194 Fab and Transmissible Mutant H5 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HAEMAGGLUTININ HA1 FAB HEAVY CHAIN, ... | | Authors: | Xiong, X, Corti, D, Liu, J, Pinna, D, Foglierini, M, Calder, L.J, Martin, S.R, Lin, Y.P, Walker, P.A, Collins, P.J, Monne, I, Suguitan Jr, A.L, Santos, C, Temperton, N.J, Subbarao, K, Lanzavecchia, A, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Complexes Formed by H5 Influenza Hemagglutinin with a Potent Broadly Neutralizing Human Monoclonal Antibody.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3CL0
 
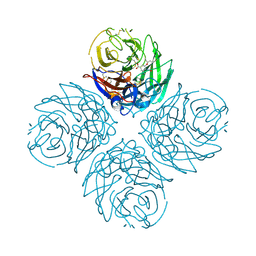 | | N1 Neuraminidase H274Y + oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Collins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
3DHE
 
 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED DEHYDROEPIANDROSTERONE | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Han, Q, Campbell, R.L, Gangloff, A, Lin, S.X. | | Deposit date: | 1998-03-25 | | Release date: | 1999-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
J.Biol.Chem., 275, 2000
|
|
2G7C
 
 | | Clostridium difficile Toxin A Fragment Bound to aGal(1,3)bGal(1,4)bGlcNAc | | Descriptor: | GLYCEROL, Toxin A, alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Greco, A, Ho, J.G.S, Lin, S.J, Palcic, M.M, Rupnik, M, Ng, K.K.S. | | Deposit date: | 2006-02-28 | | Release date: | 2006-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate recognition by Clostridium difficile toxin A.
Nat.Struct.Mol.Biol., 13, 2006
|
|
8XP9
 
 | |
8XPB
 
 | | Crystal structure of d(ACGCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Huang, H.T, Lin, S.M, Neidle, S. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of d(ACGCCGT/ACGGCGT) in complex with Echinomycin
To Be Published
|
|
8WR4
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
