8AHQ
 
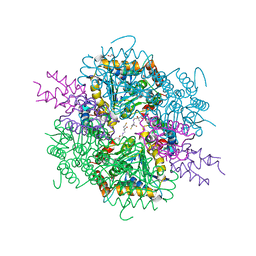 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AHZ
 
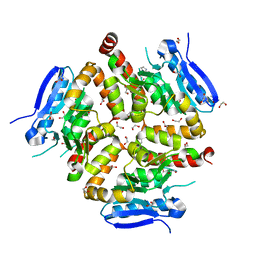 | | Native VirD of Streptomyces virginiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enoyl-CoA hydratase, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
4D6X
 
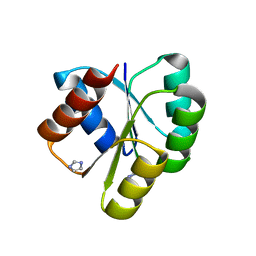 | | Crystal structure of the receiver domain of NtrX from Brucella abortus | | Descriptor: | BACTERIAL REGULATORY, FIS FAMILY PROTEIN, IMIDAZOLE | | Authors: | Klinke, S, Fernandez, I, Carrica, M.C, Otero, L.H, Goldbaum, F.A. | | Deposit date: | 2014-11-18 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Snapshots of Conformational Changes Shed Light Into the Ntrx Receiver Domain Signal Transduction Mechanism
J.Mol.Biol., 427, 2015
|
|
8AIG
 
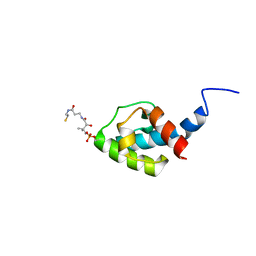 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8ALL
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
1MUC
 
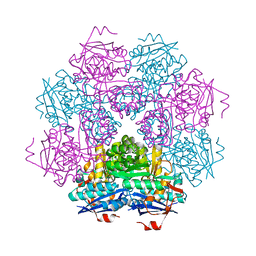 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
3OLL
 
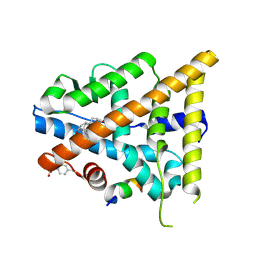 | | Crystal structure of phosphorylated estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
3OMO
 
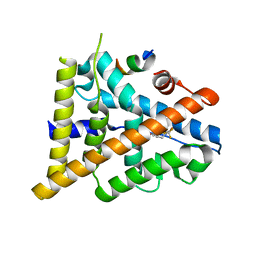 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-(trifluoroacetyl)-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
4ER4
 
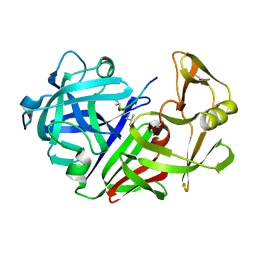 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
3OMQ
 
 | | Fragment-Based Design of novel Estrogen Receptor Ligands | | Descriptor: | 2-[(trifluoromethyl)sulfonyl]-1,2,3,4-tetrahydroisoquinolin-6-ol, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, van Otterlo, W.A, Rose, R, Fuchs, S, Dominguez Seoane, M, Waldmann, H, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Evaluation of Fragment-Like Estrogen Receptor Tetrahydroisoquinoline Ligands from a Scaffold-Detection Approach.
J.Med.Chem., 54, 2011
|
|
6UGX
 
 | | Crystal structure of the Fc fragment of PF06438179/GP1111 an infliximab biosimilar in a primative orthorhombic crystal form, Lot A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
6UGY
 
 | | Crystal structure of the Fc fragment of anti-TNFa antibody infliximab (Remicade) in a primative orthorhombic crystal form, Lot C | | Descriptor: | ACETATE ION, Remicade Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6HQP
 
 | | Crystal structure of GcoA F169V bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5VH5
 
 | | Crystal Structure of Fc fragment of anti-TNFa antibody infliximab | | Descriptor: | ACETATE ION, Infliximab Fc, ZINC ION, ... | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
5VH3
 
 | | Crystal structure of Fab fragment of the anti-TNFa antibody infliximab in a C-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
6HQS
 
 | | Crystal structure of GcoA F169S bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5VH4
 
 | | Crystal structure of Fab fragment of anti-TNFa antibody infliximab in an I-centered orthorhombic crystal form | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Mayclin, S.J, Edwards, T.E, Lerch, T.F, Conlan, H, Sharpe, P. | | Deposit date: | 2017-04-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Infliximab crystal structures reveal insights into self-association.
MAbs, 9, 2017
|
|
6HQQ
 
 | | Crystal structure of GcoA F169A bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4L8L
 
 | |
6HQL
 
 | | Crystal structure of GcoA F169H bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQT
 
 | | Crystal structure of GcoA F169V bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQO
 
 | | Crystal structure of GcoA F169S bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQN
 
 | | Crystal structure of GcoA F169L bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4LIK
 
 | |
