1B2I
 
 | |
1ZMY
 
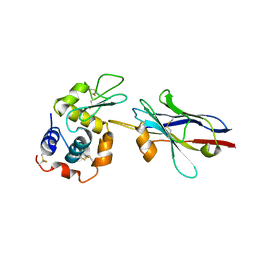 | | cAbBCII-10 VHH framework with CDR loops of cAbLys3 grafted on it and in complex with hen egg white lysozyme | | Descriptor: | Antibody cabbcII-10:lys3, Lysozyme C | | Authors: | Saerens, D, Pellis, M, Loris, R, Pardon, E, Dumoulin, M, Matagne, A, Wyns, L, Muyldermans, S, Conrath, K. | | Deposit date: | 2005-05-11 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies
J.Mol.Biol., 352, 2005
|
|
1MDG
 
 | | An Alternating Antiparallel Octaplex in an RNA Crystal Structure | | Descriptor: | 5'-R(*UP*(BGM)GP*AP*GP*GP*U)-3', COBALT HEXAMMINE(III), SODIUM ION | | Authors: | Pan, B.C, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction
Structure, 11, 2003
|
|
1J8G
 
 | | X-ray Analysis of a RNA Tetraplex r(uggggu)4 at Ultra-High Resolution | | Descriptor: | 5'-R(*UP*GP*GP*GP*GP*U)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Deng, J, Xiong, Y, Sundaralingam, M. | | Deposit date: | 2001-05-21 | | Release date: | 2001-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.61 Å) | | Cite: | X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 A).
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
438D
 
 | |
1L3Z
 
 | | Crystal Structure Analysis of an RNA Heptamer | | Descriptor: | 5'-R(*GP*UP*AP*UP*AP*CP*A)-3', SODIUM ION | | Authors: | Shi, K, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-03-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of an alternating RNA heptamer r(GUAUACA)
forming a six base-paired duplex with 3'-end adenine overhangs
Nucleic Acids Res., 31, 2003
|
|
1J9H
 
 | | Crystal Structure of an RNA Duplex with Uridine Bulges | | Descriptor: | 5'-R(*GP*UP*GP*UP*CP*GP*(CBR)P*AP*C)-3', CALCIUM ION | | Authors: | Xiong, Y, Deng, J, Sudarsanakumar, C, Sundaralingam, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA duplex r(gugucgcac)(2) with uridine bulges.
J.Mol.Biol., 313, 2001
|
|
1I6Z
 
 | | BAG DOMAIN OF BAG1 COCHAPERONE | | Descriptor: | BAG-FAMILY MOLECULAR CHAPERONE REGULATOR-1 | | Authors: | Briknarova, K, Takayama, S, Brive, L, Havert, M.L, Knee, D.A, Velasco, J, Homma, S, Cabezas, E, Stuart, J, Hoyt, D.W, Satterthwait, A.C, Llinas, M, Reed, J.C, Ely, K.R. | | Deposit date: | 2001-03-06 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein.
Nat.Struct.Biol., 8, 2001
|
|
4CML
 
 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CXL
 
 | | Human insulin analogue (D-ProB8)-insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
1M6R
 
 | |
2XT6
 
 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
4CXN
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form I | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
4BCT
 
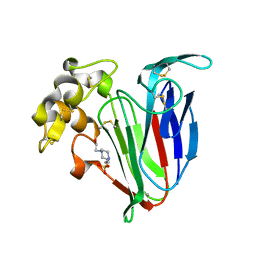 | | Crystal structure of kiwi-fruit allergen Act d 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, THAUMATIN-LIKE PROTEIN | | Authors: | Pavkov-Keller, T, Bublin, M, Jankovic, M, Breiteneder, H, Keller, W. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structure of Kiwi-Fruit Allergen Act D 2
To be Published
|
|
2YIC
 
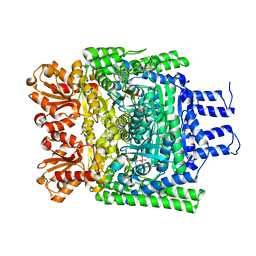 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
1JO2
 
 | |
2XTA
 
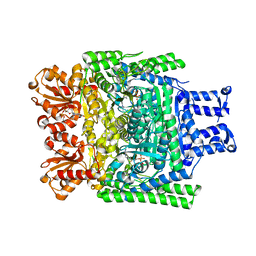 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with acetyl-CoA (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
3BIV
 
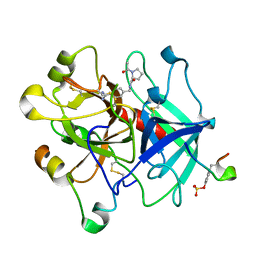 | | Human thrombin-in complex with UB-THR11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
4CY7
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form II | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
305D
 
 | |
306D
 
 | |
2Y0P
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate and acetyl-CoA | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, ACETYL COENZYME *A, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
2YID
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase in complex with the enamine-ThDP intermediate | | Descriptor: | (4E)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-2(3H)-ylidene}-4-hydroxybutanoic acid, 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
4BJ0
 
 | | Xyloglucan binding module (CBM4-2 X2-L110F) in complex with branched xyloses | | Descriptor: | CALCIUM ION, XYLANASE, alpha-D-glucopyranose, ... | | Authors: | Schantz, L, Hakansson, M, Logan, D.T, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2013-04-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carbohydrate Binding Module Recognition of Xyloglucan Defined by Polar Contacts with Branching Xyloses and Ch-Pi Interactions.
Proteins, 82, 2014
|
|
