7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
5LAE
 
 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
6NCA
 
 | | HLA-A2 (A*02:01) bound to a peptide from the Epstein-Barr virus BRLF1 protein | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Stern, L.J, Selin, L.K, Song, I.Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (3.300001 Å) | | Cite: | CDR3 alpha drives selection of the immunodominant Epstein Barr virus (EBV) BRLF1-specific CD8 T cell receptor repertoire in primary infection.
Plos Pathog., 15, 2019
|
|
6H6S
 
 | | Sad phasing on nickel-substituted human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, NICKEL (II) ION | | Authors: | Calderone, V, Fragai, M, Silva, J.P, Luchinat, C, Ravera, E, Geraldes, C.F.G.C, Macedo, A.L, Cerofolini, L, Giuntini, S. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-crystallographic symmetry in proteins: Jahn-Teller-like and Butterfly-like effects?
J. Biol. Inorg. Chem., 24, 2019
|
|
6U1A
 
 | | Crystal Structure of Fluorescent Protein FusionRed | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Red fluorescent protein | | Authors: | Pletnev, S, Muslinkina, L, Pletneva, N, Pletnev, V.Z. | | Deposit date: | 2019-08-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Two independent routes of post-translational chemistry in fluorescent protein FusionRed.
Int.J.Biol.Macromol., 155, 2020
|
|
6USC
 
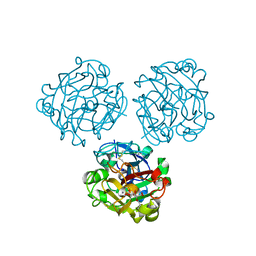 | | Structure of Human Intelectin-1 in complex with KO | | Descriptor: | CALCIUM ION, CHLORIDE ION, Intelectin-1, ... | | Authors: | Windsor, I.W, Isabella, C.R, Kosma, P, Raines, R.T, Kiessling, L.L. | | Deposit date: | 2019-10-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Stereoelectronic Effects Impact Glycan Recognition.
J.Am.Chem.Soc., 142, 2020
|
|
6VAH
 
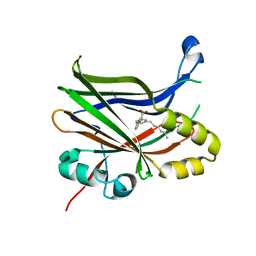 | | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative | | Descriptor: | 2-fluoro-6-[(3-hexylphenyl)amino]benzoic acid, Transcriptional enhancer factor TEF-4, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Dong, A, Li, Y, Melin, L, Gagnon, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human TEAD2 transcription factor in complex with Flufenamic acid derivative
to be published
|
|
6MII
 
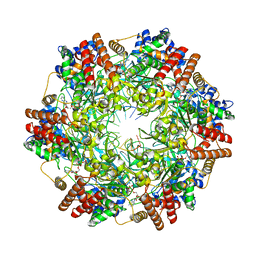 | | Crystal structure of minichromosome maintenance protein MCM/DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Meagher, M, Epling, L.B. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | DNA translocation mechanism of the MCM complex and implications for replication initiation.
Nat Commun, 10, 2019
|
|
6N99
 
 | | Xylose isomerase 2F1 variant from Streptomyces sp. F-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6YU6
 
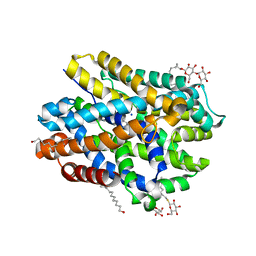 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU4
 
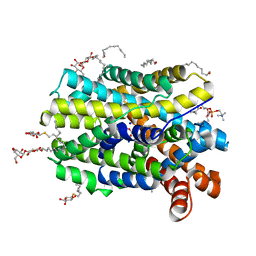 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
7PRM
 
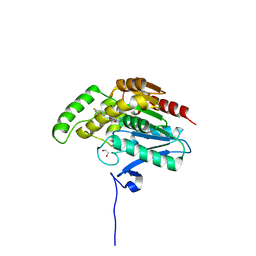 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 13 | | Descriptor: | (4~{R})-1-[4-(4-fluorophenyl)phenyl]-4-[4-(furan-2-ylcarbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Grether, U, Gobbi, L, Kuhn, B, Collin, L, Leibrock, L, Heer, D, Wittwer, M, Benz, J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of High Brain-Penetrant and Reversible Monoacylglycerol Lipase PET Tracers for Neuroimaging.
J.Med.Chem., 65, 2022
|
|
6N98
 
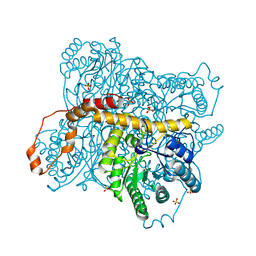 | | Xylose isomerase 1F1 variant from Streptomyces sp. F-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, Xylose isomerase | | Authors: | Miyamoto, R.Y, Vieira, P.S, Murakami, M.T, Zanphorlin, L.M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a novel xylose isomerase from Streptomyces sp. F-1 revealed the presence of unique features that differ from conventional classes.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6YS6
 
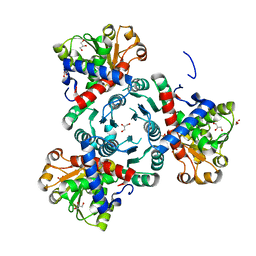 | | Arabidopsis aspartate transcarbamoylase complex with PALA | | Descriptor: | GLYCEROL, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, PYRB, ... | | Authors: | Ramon Maiques, S, Del Cano Ochoa, F, Bellin, L, Mohlmann, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanisms of feedback inhibition and sequential firing of active sites in plant aspartate transcarbamoylase.
Nat Commun, 12, 2021
|
|
6XTU
 
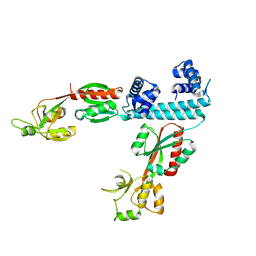 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XTV
 
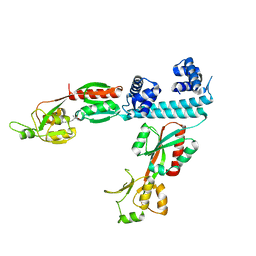 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6YU5
 
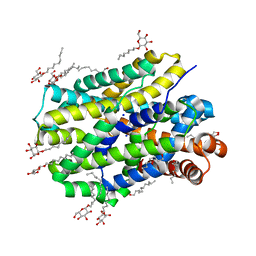 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6ZFC
 
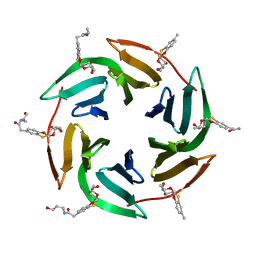 | | Fucose-binding lectin from Burkholderia ambifaria (BamBL) in complex with a fucosyl derivative | | Descriptor: | 2-[(2~{S},3~{S},4~{R},4~{a}~{S},10~{a}~{S})-2-methyl-3,4-bis(oxidanyl)-3,4,4~{a},10~{a}-tetrahydro-2~{H}-pyrano[2,3-b][1,4]benzoxathiin-7-yl]-~{N}-(3-oxidanylpropyl)ethanamide, bacterial lectin from Burkholderia ambifaria | | Authors: | Kuhaudomlarp, S, Gillon, E, Fragai, M, Cerofolini, L, Giuntini, S, Denis, M, Santarsia, S, Valori, C, Dondoni, A, Fallarini, S, Lombardi, G, Nativi, C, Imberty, A. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fucosylated ubiquitin and orthogonally glycosylated mutant A28C: conceptually new ligands for Burkholderia ambifaria lectin (BambL).
Chem Sci, 11, 2020
|
|
6YU7
 
 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU3
 
 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
8S8I
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8D
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8G
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8H
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
