4DHU
 
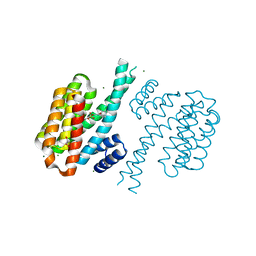 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(2,3-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4HW1
 
 | | Multiple Crystal structures of an all-AT DNA dodecamer stabilized by weak interactions. | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3'), MAGNESIUM ION | | Authors: | Acosta-Reyes, F, Subirana, J.A, Pous, J, Condom, N, Malinina, L, Campos, J.L. | | Deposit date: | 2012-11-07 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
4DHM
 
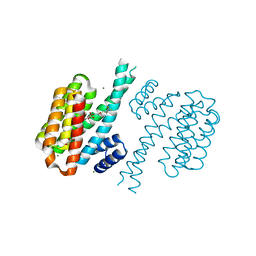 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4DHT
 
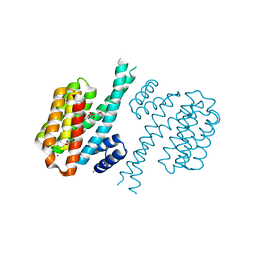 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | 14-3-3 PROTEIN SIGMA, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
4GXD
 
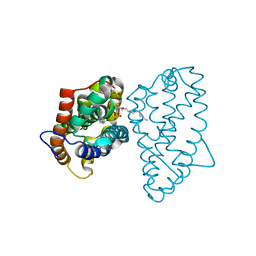 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1AZN
 
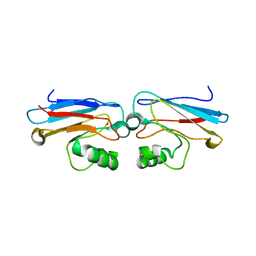 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT PHE114ALA FROM PSEUDOMONAS AERUGINOSA AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Pascher, T, Nar, H. | | Deposit date: | 1994-05-27 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the azurin mutant Phe114Ala from Pseudomonas aeruginosa at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
4H2Z
 
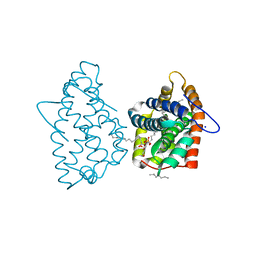 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | Authors: | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | Deposit date: | 2012-09-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3RZN
 
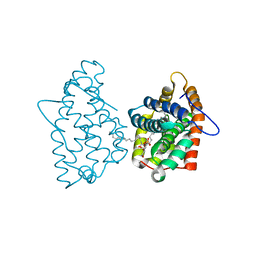 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
1GR7
 
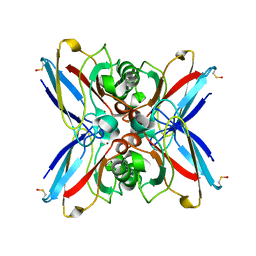 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
3SHI
 
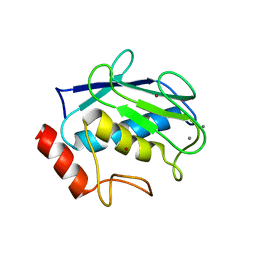 | | Crystal structure of human MMP1 catalytic domain at 2.2 A resolution | | Descriptor: | CALCIUM ION, Interstitial collagenase, ZINC ION | | Authors: | Bertini, I, Calderone, V, Cerofolini, L, Fragai, M, Geraldes, C.F.G.C, Hermann, P, Luchinat, C, Parigi, G, Teixeira, J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of MMP-1 studied through tagged lanthanides.
Febs Lett., 586, 2012
|
|
2PT0
 
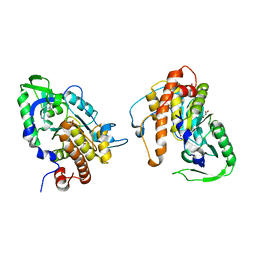 | |
2PSZ
 
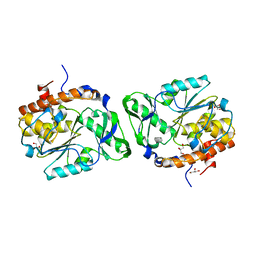 | |
1OOW
 
 | | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f | | Descriptor: | COPPER (II) ION, Plastocyanin, chloroplast | | Authors: | Jansson, H, Okvist, M, Jacobson, F, Ejdeback, M, Hansson, O, Sjolin, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the spinach plastocyanin double mutant G8D/L12E gives insight into its low reactivity towards photosystem 1 and cytochrome f.
Biochim.Biophys.Acta, 1607, 2003
|
|
2QMI
 
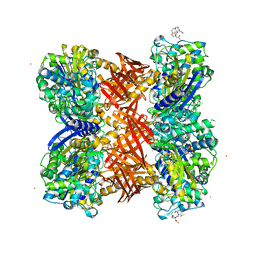 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
1N70
 
 | |
1A7C
 
 | | HUMAN PLASMINOGEN ACTIVATOR INHIBITOR TYPE-1 IN COMPLEX WITH A PENTAPEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-ribopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAPEPTIDE, ... | | Authors: | Xue, Y, Inghardt, T, Sjolin, L, Deinum, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interfering with the inhibitory mechanism of serpins: crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactive-centre loop peptide
Structure, 6, 1998
|
|
1P8L
 
 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
1FN2
 
 | | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
1BY3
 
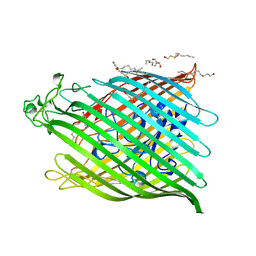 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1C0A
 
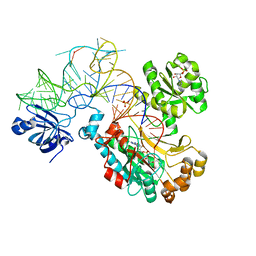 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
2VNV
 
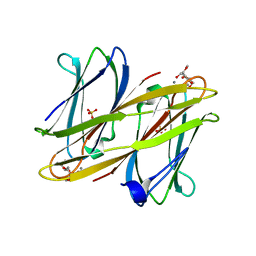 | | Crystal structure of BclA lectin from burkholderia cenocepacia in complex with alpha-methyl-mannoside at 1.7 Angstrom resolution | | Descriptor: | BCLA, CALCIUM ION, SULFATE ION, ... | | Authors: | Lameignere, E, Malinovska, L, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Mannose Recognition by a Lectin from Opportunistic Bacteria Burkholderia Cenocepacia
Biochem.J., 411, 2008
|
|
1COB
 
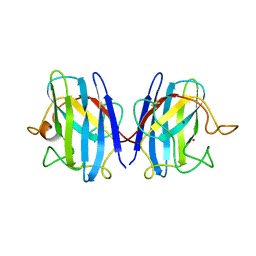 | | CRYSTAL STRUCTURE SOLUTION AND REFINEMENT OF THE SEMISYNTHETIC COBALT SUBSTITUTED BOVINE ERYTHROCYTE ENZYME SUPEROXIDE DISMUTASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | COBALT (II) ION, COPPER (II) ION, SUPEROXIDE DISMUTASE | | Authors: | Djinovic, K, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Rotilio, G, Bolognesi, M. | | Deposit date: | 1992-02-19 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure solution and refinement of the semisynthetic cobalt-substituted bovine erythrocyte superoxide dismutase at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1FN1
 
 | | CRYSTAL STRUCTURE OF 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CG(5BR)UACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
1MZY
 
 | | Crystal Structure of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MAGNESIUM ION | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
1B8A
 
 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
