7OSQ
 
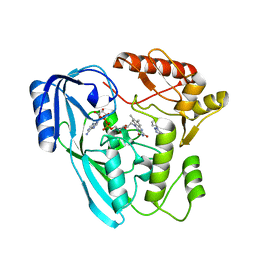 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
4RW2
 
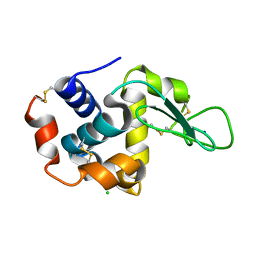 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: refocused beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Barends, T, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
4WN4
 
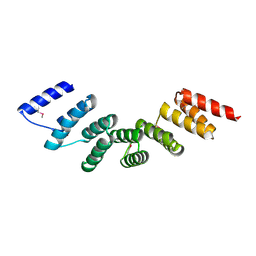 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
7PP7
 
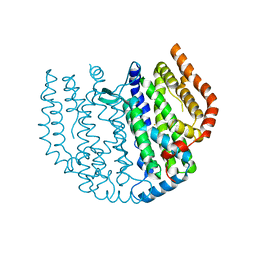 | | Thunberia alata 16:0-ACP desaturase | | Descriptor: | Acyl-[acyl-carrier-protein] 6-desaturase, FE (III) ION | | Authors: | Guy, J.E, Whittle, E, Cai, Y, Chai, J, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2021-09-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselectivity mechanism of the Thunbergia alata Delta 6-16:0-acyl carrier protein desaturase.
Plant Physiol., 188, 2022
|
|
4WSL
 
 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
6I48
 
 | | Structure of P. aeruginosa LpxC with compound 12: (2R)-4-(6-(2-Fluoro-4-methoxyphenyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, 1,2-ETHANEDIOL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
4RW1
 
 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: original beam | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Foucar, L, Botha, S, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | Deposit date: | 2014-12-01 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
4U7W
 
 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
4U33
 
 | | Structure of Mtb GlgE bound to maltose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4TWE
 
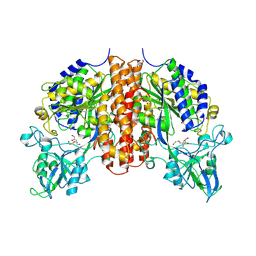 | | Structure of ligand-free N-acetylated-alpha-linked-acidic-dipeptidase like protein (NAALADaseL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Barinka, C, Lubkowski, J, Sacha, P, Konvalinka, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of a novel aminopeptidase from human intestine.
J.Biol.Chem., 290, 2015
|
|
5LCB
 
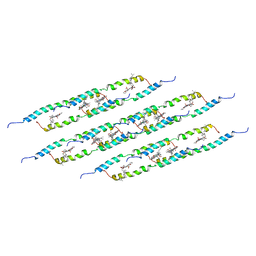 | | In situ atomic-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum obtained combining solid-state NMR spectroscopy, cryo electron microscopy and polarization spectroscopy | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll c-binding protein | | Authors: | Nielsen, J.T, Kulminskaya, N.V, Bjerring, M, Linnanto, J.M, Ratsep, M, Pedersen, M, Lambrev, P.H, Dorogi, M, Garab, G, Thomsen, K, Jegerschold, C, Frigaard, N.U, Lindahl, M, Nielsen, N.C. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (26.5 Å), SOLID-STATE NMR | | Cite: | In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum.
Nat Commun, 7, 2016
|
|
4UMQ
 
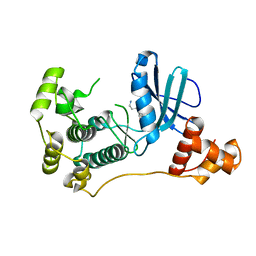 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-{5-[(3-hydroxy-5-methoxyphenyl)amino]-2-(phenylcarbamoyl)phenoxy}propan-1-aminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMT
 
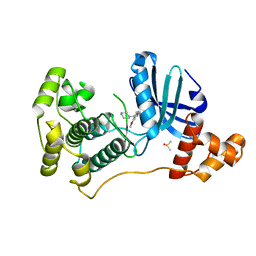 | | Structure of MELK in complex with inhibitors | | Descriptor: | 1-(4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}-2-methoxybenzyl)piperazinediium, DIMETHYL SULFOXIDE, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMU
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | (2-ethoxy-4-{[3-(isoquinolin-7-yl)prop-2-yn-1-yl]oxy}phenyl)methanaminium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase.
Acs Med.Chem.Lett., 6, 2015
|
|
4UMR
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4U3C
 
 | | Docking Site of Maltohexaose in the Mtb GlgE | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U2Y
 
 | | Sco GlgEI-V279S in Complex with Reaction Intermediate Azasugar | | Descriptor: | (2R,3R,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-3-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1 | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U31
 
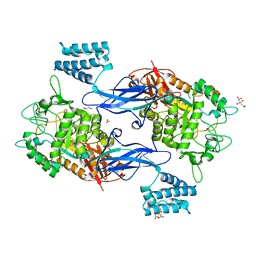 | | Sco GlgEI-V279S in Complex with maltose-C-phosphonate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U2Z
 
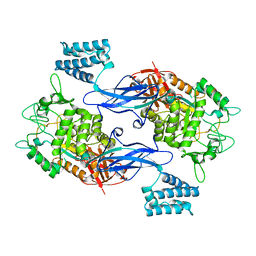 | | X-ray crystal structure of an Sco GlgEI-V279S/1,2,2-trifluromaltose complex | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2,2-difluoro-alpha-D-arabino-hexopyranosyl fluoride | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis of 2-deoxy-2,2-difluoro-alpha-maltosyl fluoride and its X-ray structure in complex with Streptomyces coelicolor GlgEI-V279S.
Org.Biomol.Chem., 13, 2015
|
|
6I47
 
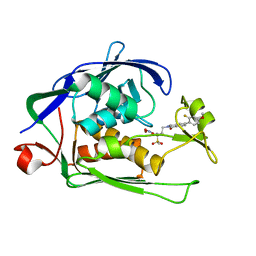 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I49
 
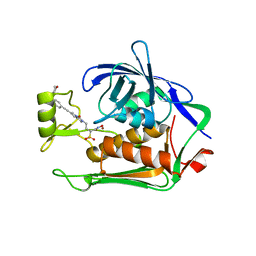 | | Structure of P. aeruginosa LpxC with compound 17a: (2R)-N-Hydroxy-2-methyl-2-(methylsulfonyl)-4(6((4(morpholinomethyl)phenyl)ethynyl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)yl)butanamide | | Descriptor: | (2~{R})-2-methyl-2-methylsulfonyl-4-[6-[2-[4-(morpholin-4-ylmethyl)phenyl]ethynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I4A
 
 | | Structure of P. aeruginosa LpxC with compound 18d: (2R)-N-Hydroxy-4-(6-((1-(hydroxymethyl)cyclopropyl)buta-1,3-diyn-1-yl)-3-oxo-1H-pyrrolo[1,2-c]imidazol-2(3H)-yl)-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-[4-[1-(hydroxymethyl)cyclopropyl]buta-1,3-diynyl]-3-oxidanylidene-1~{H}-pyrrolo[1,2-c]imidazol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6I46
 
 | | Structure of P. aeruginosa LpxC with compound 8: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-2-oxooxazol-3(2H)-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[5-(2-fluoranyl-4-methoxy-phenyl)-2-oxidanylidene-1,3-oxazol-3-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, GLYCEROL, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
4QYX
 
 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
