7UCX
 
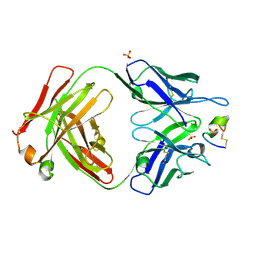 | | LRP8 11H1 Fab complexed to a cyclized CR1 peptide | | Descriptor: | 11H1 Fab Heavy chain, 11H1 Fab Light chain, Cyclized CR1 peptide, ... | | Authors: | Argiriadi, M.A, Deng, K, Egan, D, Gao, L, Gizatullin, F, Harlan, J, Karaoglu, D, Qiu, W, Goodearl, A. | | Deposit date: | 2022-03-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The use of cyclic peptide antigens to generate LRP8 specific antibodies
Front Drug Discov (Lausanne), 2, 2023
|
|
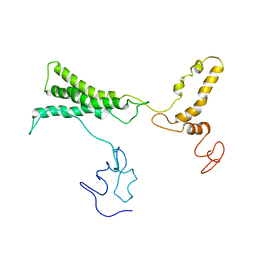
8TVL
 
 | |
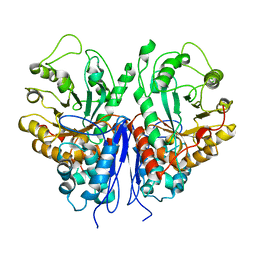
8UOP
 
 | |
8UOY
 
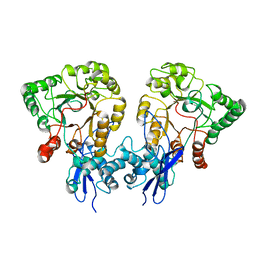 | |
7UGU
 
 | |
6WY6
 
 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | Descriptor: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | Authors: | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
5HPG
 
 | |
1UG3
 
 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
6UZ4
 
 | |
6UZ5
 
 | |
5V4U
 
 | |
1I5K
 
 | | STRUCTURE AND BINDING DETERMINANTS OF THE RECOMBINANT KRINGLE-2 DOMAIN OF HUMAN PLASMINOGEN TO AN INTERNAL PEPTIDE FROM A GROUP A STREPTOCOCCAL SURFACE PROTEIN | | Descriptor: | M PROTEIN, PLASMINOGEN | | Authors: | Rios-Steiner, J.L, Schenone, M, Mochalkin, I, Tulinsky, A, Castellino, F.J. | | Deposit date: | 2001-02-27 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein.
J.Mol.Biol., 308, 2001
|
|
1QIC
 
 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
1QIA
 
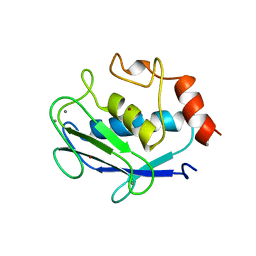 | | CRYSTAL STRUCTURE OF STROMELYSIN CATALYTIC DOMAIN | | Descriptor: | CALCIUM ION, STROMELYSIN-1, ZINC ION | | Authors: | Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A.G, Rubin, J.R, Skeean, R.W, White, A.D, Blundell, T.L, Humblet, C, Hupe, D.J, Dhanaraj, V. | | Deposit date: | 1999-06-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity
Protein Sci., 8, 1999
|
|
5TGL
 
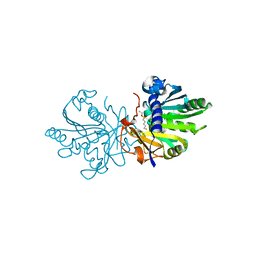 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
2N7O
 
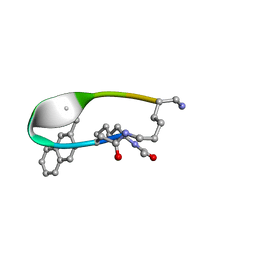 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
1QIB
 
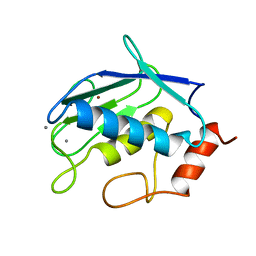 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
6BZL
 
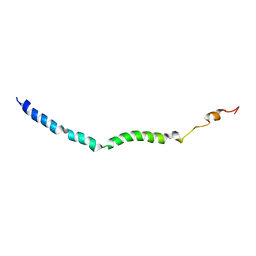 | | Solution structure of VEK75 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
4JC2
 
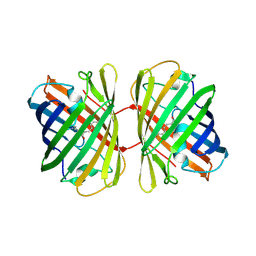 | | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral | | Descriptor: | asFP504 | | Authors: | Huang, Y.C, Emralino, F.L, Liu, F.C, Saloma, C.P, Bascos, N.A, Chen, C.J. | | Deposit date: | 2013-02-21 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral
To be Published
|
|
1Y0J
 
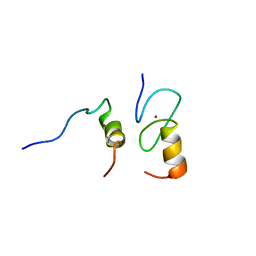 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2N7T
 
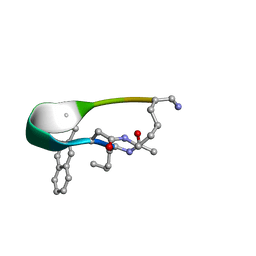 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7N
 
 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
6BZJ
 
 | | Solution structure of AGL55 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
1KI0
 
 | | The X-ray Structure of Human Angiostatin | | Descriptor: | ANGIOSTATIN, BICINE | | Authors: | Abad, M.C, Arni, R.K, Grella, D.K, Castellino, F.J, Tulinsky, A, Geiger, J.H. | | Deposit date: | 2001-12-02 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin.
J.Mol.Biol., 318, 2002
|
|
3H4V
 
 | | Selective screening and design to identify inhibitors of leishmania major pteridine reductase 1 | | Descriptor: | METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL]AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Mcluskey, K, Gibellini, F, Hunter, W.N. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
