2MKO
 
 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
1J91
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the ATP-competitive inhibitor 4,5,6,7-tetrabromobenzotriazole | | Descriptor: | 4,5,6,7-TETRABROMOBENZOTRIAZOLE, CASEIN KINASE II, ALPHA CHAIN | | Authors: | Battistutta, R, De Moliner, E, Sarno, S, Zanotti, G, Pinna, L.A. | | Deposit date: | 2001-05-23 | | Release date: | 2002-05-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole.
Protein Sci., 10, 2001
|
|
4ALO
 
 | | STRUCTURE AND PROPERTIES OF H1 CRUSTACYANIN FROM LOBSTER HOMARUS AMERICANUS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, H1 APOCRUSTACYANIN, SODIUM ION, ... | | Authors: | Ferrari, M, Folli, C, Pincolini, E, Mcclintock, T.S, Roessle, M, Berni, R, Cianci, M. | | Deposit date: | 2012-03-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Recombinant Crustacyanin Subunits from the Lobster Homarus Americanus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2JCC
 
 | | AH3 recognition of mutant HLA-A2 W167A | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ... | | Authors: | Miller, P, Benhar, Y.P, Biddison, W, Collins, E.J. | | Deposit date: | 2006-12-21 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Single Mhc Mutation Eliminates Enthalpy Associated with T Cell Receptor Binding.
J.Mol.Biol., 373, 2007
|
|
1IA0
 
 | | KIF1A HEAD-MICROTUBULE COMPLEX STRUCTURE IN ATP-FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, ... | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-03-22 | | Release date: | 2002-03-22 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Switch-based Mechanism of Kinesin Motors
Nature, 411, 2001
|
|
4BTX
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-isopropylamino-2-phenyl-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, CALCIUM ION, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
2N7O
 
 | | NMR Structure of Peptide PG-990 in DPC micelles | | Descriptor: | Peptide PG-990 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
4BTW
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(cyclohexylamino)-2-phenyl-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
2N7N
 
 | | NMR structure of Peptide PG-989 in DPC micelles | | Descriptor: | Peptide PG-989 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-16 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2N7T
 
 | | NMR structure of Peptide PG-992 in DPC micelles | | Descriptor: | Peptide PG-992 | | Authors: | Carotenuto, A, Merlino, F, Chai, M, Brancaccio, D, Yousif, A, Novellino, E, Hruby, V, Grieco, P. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2017-10-11 | | Method: | SOLUTION NMR | | Cite: | Discovery of Novel Potent and Selective Agonists at the Melanocortin-3 Receptor.
J.Med.Chem., 58, 2015
|
|
2C86
 
 | | x-ray structure of the N and C-terminal domain of coronavirus nucleocapsid protein. | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collinson, E.W, Lescar, J, Prasad, B.V.V. | | Deposit date: | 2005-12-02 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structures of the N- and C-Terminal Domains of a Coronavirus Nucleocapsid Protein: Implications for Nucleocapsid Formation.
J.Virol., 80, 2006
|
|
4BTY
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-(4-methylpiperazin-1-yl)phenylamino]-2-(4-chlorophenyl)-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, ... | | Authors: | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
2B1P
 
 | | inhibitor complex of JNK3 | | Descriptor: | 3-{6-[(2-CHLOROPHENYL)AMINO]-1H-INDAZOL-3-YL}-5-{[4-(DIMETHYLAMINO)BUTANOYL]AMINO}BENZOIC ACID, BETA-MERCAPTOETHANOL, Mitogen-activated protein kinase 10, ... | | Authors: | Swahn, B.M, Huerta, F, Kallin, E, Malmstrom, J, Weigelt, T, Viklund, J, Womack, P, Xue, Y, Ohberg, L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2VVO
 
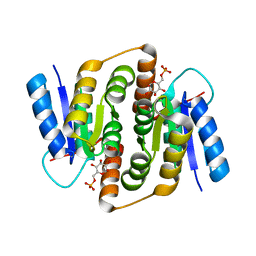 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with alpha d-allose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-allopyranose, RIBOSE-5-PHOSPHATE ISOMERASE B | | Authors: | Roos, A.K, Mariano, S, Kowalinski, E, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
2KVY
 
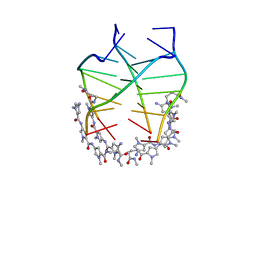 | | NMR solution structure of the 4:1 complex between an uncharged distamycin A analogue and [d(TGGGGT)]4 | | Descriptor: | 4-amino-1-methyl-N-{1-methyl-5-[(1-methyl-5-{[3-(methylamino)-3-oxopropyl]carbamoyl}-1H-pyrrol-3-yl)carbamoyl]-1H-pyrrol-3-yl}-1H-pyrrole-2-carboxamide, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3') | | Authors: | Cosconati, S, Marinelli, L, Trotta, R, Virno, A, De Tito, S, Romagnoli, R, Pagano, B, Limongelli, V, Giancola, C, Baraldi, P, Mayol, L, Novellino, E, Randazzo, A. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle.
J.Am.Chem.Soc., 132, 2010
|
|
1I5S
 
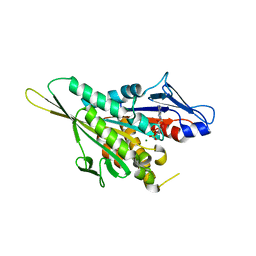 | | CRYSTAL STRUCTURE OF THE KIF1A MOTOR DOMAIN COMPLEXED WITH MG-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF1A, MAGNESIUM ION | | Authors: | Kikkawa, M, Sablin, E.P, Okada, Y, Yajima, H, Fletterick, R.J, Hirokawa, N. | | Deposit date: | 2001-02-28 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Switch-based mechanism of kinesin motors
Nature, 411, 2001
|
|
4KZC
 
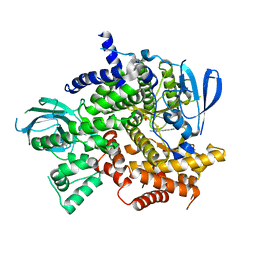 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ZQT
 
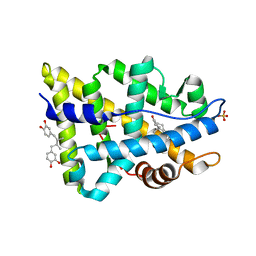 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
1IMW
 
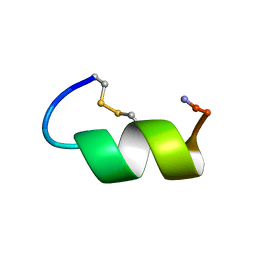 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1Z6C
 
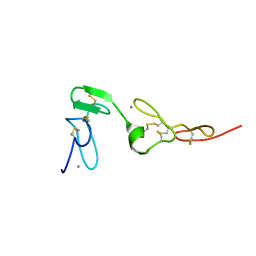 | | Solution structure of an EGF pair (EGF34) from vitamin K-dependent protein S | | Descriptor: | CALCIUM ION, Vitamin K-dependent protein S | | Authors: | Drakenberg, T, Ghasriani, H, Thulin, E, Thamlitz, A.M, Muranyi, A, Annila, A, Stenflo, J. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ca(2+)-Binding EGF3-4 Pair from Vitamin K-Dependent Protein S: Identification of an Unusual Fold in EGF3.
Biochemistry, 44, 2005
|
|
1JAM
 
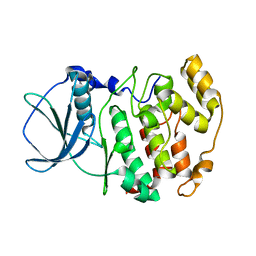 | | Crystal structure of apo-form of Z. Mays CK2 protein kinase alpha subunit | | Descriptor: | CASEIN KINASE II, ALPHA CHAIN | | Authors: | Battistutta, R, De Moliner, E, Sarno, S, Zanotti, G, Pinna, L.A. | | Deposit date: | 2001-05-31 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole.
Protein Sci., 10, 2001
|
|
2LV3
 
 | |
2NZZ
 
 | | NMR structure analysis of the Penetratin conjugated Gas (374-394) peptide | | Descriptor: | Penetratin conjugated Gas (374-394) peptide | | Authors: | D'Ursi, A.M, Rovero, P, Albrizio, S, Espsito, C, D'Errico, G, Novellino, E. | | Deposit date: | 2006-11-27 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Driving forces in the delivery of penetratin conjugated G protein fragment.
J.Med.Chem., 50, 2007
|
|
