5Z1B
 
 | |
5Z18
 
 | |
3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3O98
 
 | | Glutathionylspermidine synthetase/amidase C59A complex with ADP and Gsp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE, ... | | Authors: | Pai, C.H, Lin, C.H, Wang, A.H.-J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
3PHJ
 
 | |
2HWR
 
 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | 2-[(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-4-YL)OXY]-2-METHYLPROPANOIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
2HWQ
 
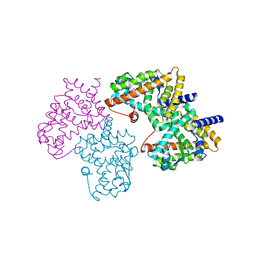 | | Structural basis for the structure-activity relationships of Peroxisome Proliferator-Activated Receptor agonists | | Descriptor: | Peroxisome proliferator-activated receptor gamma, [(1-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPYL}-1H-INDOL-5-YL)OXY]ACETIC ACID | | Authors: | Peng, Y.H, Lu, I.L, Mahindroo, N, Lin, C.H, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the structure-activity relationships of peroxisome proliferator-activated receptor agonists
J.Med.Chem., 49, 2006
|
|
3PHL
 
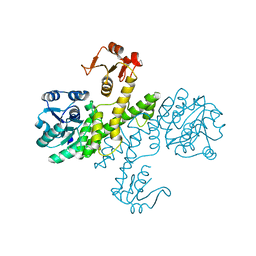 | | The apo-form UDP-glucose 6-dehydrogenase | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.Y, Ko, T.P, Lin, C.H, Chen, W.H, Wang, A.H.J. | | Deposit date: | 2010-11-04 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
1HX4
 
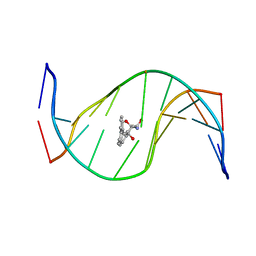 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HWV
 
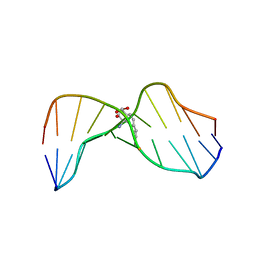 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
2IO7
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and AMPPNP | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IOB
 
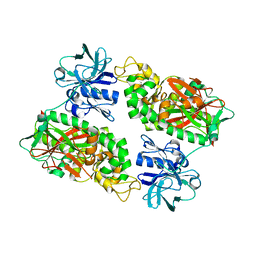 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Apo protein | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IO9
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ ,GSH and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2XII
 
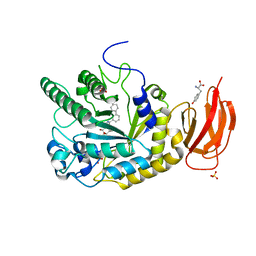 | | CRYSTAL STRUCTURE OF AN ALPHA-L-FUCOSIDASE GH29 FROM BACTEROIDES THETAIOTAOMICRON IN COMPLEX WITH AN EXTENDED 9-FLUORENONE IMINOSUGAR INHIBITOR | | Descriptor: | 9-oxo-N-[[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyl-piperidin-2-yl]methyl]fluorene-1-carboxamide, ALPHA-L-FUCOSIDASE, GLYCEROL, ... | | Authors: | Lammerts van Bueren, A, Popat, S.D, Lin, C.H, Davies, G.J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Thermodynamic Analyses of Alpha-L-Fucosidase Inhibitors.
Chembiochem, 11, 2010
|
|
2XIB
 
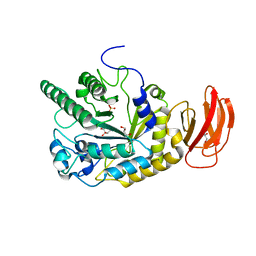 | | CRYSTAL STRUCTURE OF AN ALPHA-L-FUCOSIDASE GH29 FROM BACTEROIDES THETAIOTAOMICRON IN COMPLEX WITH DEOXYFUCONOJIRIMYCIN | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-L-FUCOSIDASE, ... | | Authors: | Lammerts van Bueren, A, Popat, S.D, Lin, C.H, Davies, G.J. | | Deposit date: | 2010-06-28 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Thermodynamic Analyses of Alpha-L-Fucosidase Inhibitors.
Chembiochem, 11, 2010
|
|
2LZK
 
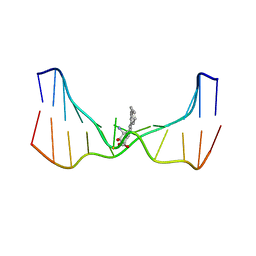 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
2MIW
 
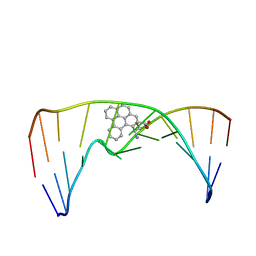 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MIV
 
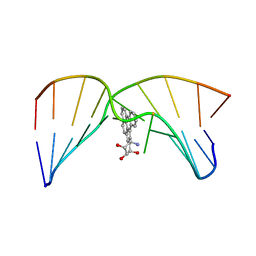 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
