3X07
 
 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
2YBQ
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6BUB
 
 | | Crystal structure of the PI3KC2alpha PX domain in space group P432 | | Descriptor: | GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
3X03
 
 | | Crystal structure of PIP4KIIBETA complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X01
 
 | | Crystal structure of PIP4KIIBETA complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X09
 
 | | Crystal structure of PIP4KIIBETA F205L complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X04
 
 | | Crystal structure of PIP4KIIBETA complex with GMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0B
 
 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
6BXH
 
 | | Menin in complex with MI-853 | | Descriptor: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Menin in complex with MI-853
To Be Published
|
|
4PQO
 
 | | Structure of the human SNX14 PX domain in space group I41 | | Descriptor: | Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
7JJC
 
 | |
4QKV
 
 | |
1TLV
 
 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
3X05
 
 | | Crystal structure of PIP4KIIBETA T201M complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X06
 
 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
2QY7
 
 | | Crystal structure of human epsinR ENTH domain | | Descriptor: | Clathrin interactor 1 | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-13 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
2C4J
 
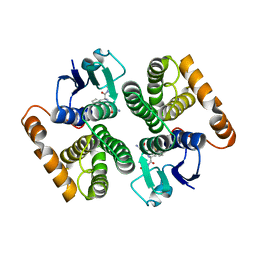 | | Human glutathione-S-transferase M2-2 T210S mutant in complex with glutathione-styrene oxide conjugate | | Descriptor: | GLUTATHIONE S-TRANSFERASE MU 2, L-GAMMA-GLUTAMYL-S-[(2S)-2-HYDROXY-2-PHENYLETHYL]-L-CYSTEINYLGLYCINE | | Authors: | Tars, K, Andersson, M, Ivarsson, Y, Olin, B, Mannervik, B. | | Deposit date: | 2005-10-20 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Alternative Mutations of a Positively Selected Residue Elicit Gain or Loss of Functionalities in Enzyme Evolution.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2R51
 
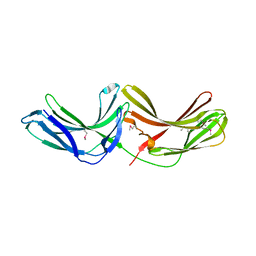 | | Crystal Structure of mouse Vps26B | | Descriptor: | Vacuolar protein sorting-associated protein 26B | | Authors: | Owen, D.J, Teasdale, R.D, Collins, B.M. | | Deposit date: | 2007-09-02 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Vps26B and mapping of its interaction with the retromer protein complex.
Traffic, 9, 2008
|
|
4HV0
 
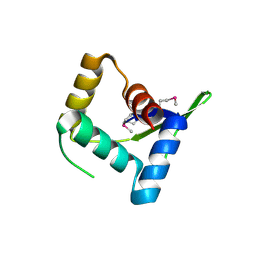 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
2C3T
 
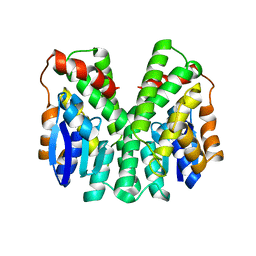 | | Human glutathione-S-transferase T1-1, W234R mutant, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1 | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-12 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
2C3N
 
 | | Human glutathione-S-transferase T1-1, apo form | | Descriptor: | GLUTATHIONE S-TRANSFERASE THETA 1, IODIDE ION | | Authors: | Tars, K, Larsson, A.-K, Shokeer, A, Olin, B, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of the Suppressed Catalytic Activity of Wild-Type Human Glutathione Transferase T1-1 Compared to its W234R Mutant.
J.Mol.Biol., 355, 2006
|
|
4MN5
 
 | | Crystal structure of PAS domain of S. aureus YycG | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Shaikh, N, Hvorup, R, Winnen, B, Collins, B.M, King, G.F. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PAS domain of S. aureus YycG
To be Published
|
|
4MN6
 
 | | Crystal structure of truncated PAS domain from S. aureus YycG | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Shaikh, N, Hvorup, R, Winnen, B, Collins, B.M, King, G.F. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PAS domain of S. aureus YycG
To be Published
|
|
1RJG
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, carboxy methyl transferase for protein phosphatase 2A catalytic subunit | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
