3X01
 
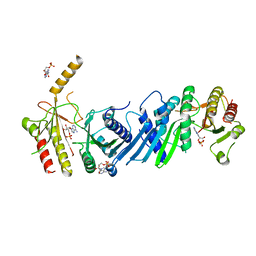 | | Crystal structure of PIP4KIIBETA complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X02
 
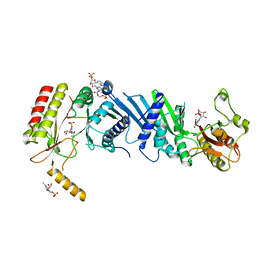 | | Crystal structure of PIP4KIIBETA complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X04
 
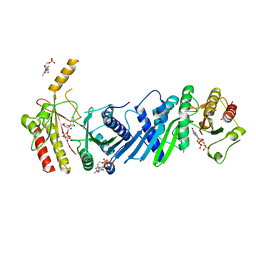 | | Crystal structure of PIP4KIIBETA complex with GMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0A
 
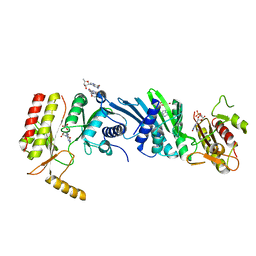 | | Crystal structure of PIP4KIIBETA F205L complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3WZZ
 
 | | Crystal structure of PIP4KIIBETA | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
1TLV
 
 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
6F8Y
 
 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine | | Descriptor: | THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
4WQ4
 
 | | E. coli YgjD(E12A)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, W, Collinet, B. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
5E7T
 
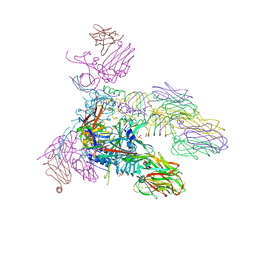 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
7ATM
 
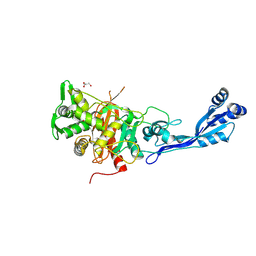 | | Structure of P. aeruginosa PBP3 in complex with a phenyl boronic acid (Compound 1) | | Descriptor: | (3-(1H-tetrazol-5-yl)phenyl)boronic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-10-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7ATO
 
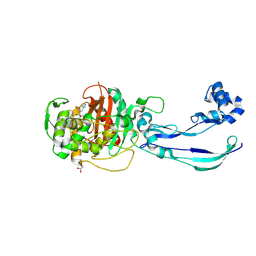 | | Structure of P. aeruginosa PBP3 in complex with an aryl boronic acid (Compound 2) | | Descriptor: | (5-methyl-1H-indazol-6-yl)boronic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-10-30 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
1RJF
 
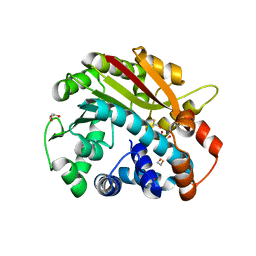 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, carboxy methyl transferase for protein phosphatase 2A catalytic subunit | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
6Q8P
 
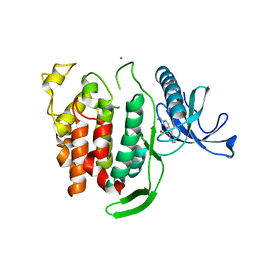 | | Structure of CLK1 with bound N-methyl-10-nitropyrido[3,4-g]quinazolin-2-amine | | Descriptor: | Dual specificity protein kinase CLK1, POTASSIUM ION, ~{N}-methyl-10-nitro-pyrido[3,4-g]quinazolin-2-amine | | Authors: | Joerger, A.C, Chatterjee, D, Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Josselin, B, Baratte, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
6Q8K
 
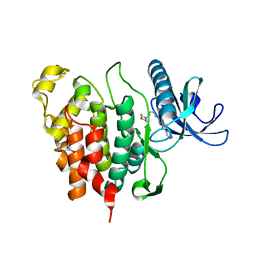 | | CLK1 with bound pyridoquinazoline | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, ~{N}2-(3-morpholin-4-ylpropyl)pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Joesselin, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
3IJE
 
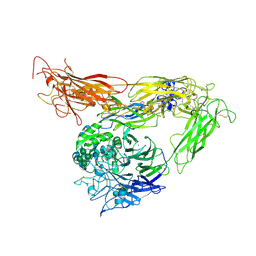 | | Crystal structure of the complete integrin alhaVbeta3 ectodomain plus an Alpha/beta transmembrane fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xiong, J.-P, Mahalingham, B, Rui, X, Hyman, B.T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment.
J.Cell Biol., 186, 2009
|
|
5OFY
 
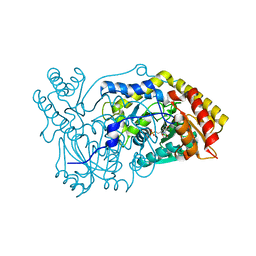 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at pH 9.0. 2.8 Ang; internal aldimine with PLP in the active site | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
5E4G
 
 | | Crystal structure of human growth differentiation factor 11 (GDF-11) | | Descriptor: | Growth/differentiation factor 11, TETRAETHYLENE GLYCOL | | Authors: | Padyana, A.K, Vaidialingam, B, Hayes, D.B, Gupta, P, Franti, M, Farrow, N.A. | | Deposit date: | 2015-10-06 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human GDF11.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5OG0
 
 | | Crystal structure of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 2.5 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
6TER
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with Galactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6F89
 
 | | Structure of H234A/Y235A P.abyssi Sua5 | | Descriptor: | BICARBONATE ION, THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
6F87
 
 | | Crystal structure of P. abyssi Sua5 complexed with L-threonine and PPi | | Descriptor: | PYROPHOSPHATE 2-, THREONINE, Threonylcarbamoyl-AMP synthase | | Authors: | Pichard-Kostuch, A, Zhang, W, Liger, D, Daugeron, M.C, Letoquart, J, Li de la Sierra-Gallay, I, Forterre, P, Collinet, B, van Tilbeurgh, H, Basta, T. | | Deposit date: | 2017-12-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t6A tRNA modification.
RNA, 24, 2018
|
|
7AO7
 
 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
1R9N
 
 | | Crystal Structure of human dipeptidyl peptidase IV in complex with a decapeptide (tNPY) at 2.3 Ang. Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Aertgeerts, K, Ye, S, Tennant, M.G, Collins, B, Rogers, J, Sang, B.-C, Skene, R, Webb, D.R, Prasad, G.S. | | Deposit date: | 2003-10-30 | | Release date: | 2005-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation.
Protein Sci., 13, 2004
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
