6GWJ
 
 | | Human OSGEP / LAGE3 / GON7 complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, EKC/KEOPS complex subunit GON7, ... | | Authors: | Missoury, S, Liger, D, Durand, D, Collinet, B, Tilbeurgh, V.H. | | Deposit date: | 2018-06-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defects in t 6 A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun, 10, 2019
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
8FMU
 
 | | Crystal structure of human Brachyury G177D variant in complex with SJF-4601 | | Descriptor: | N-(3-chloro-4-fluorophenyl)-3-[4-(dimethylamino)butanamido]-4-methoxybenzamide, T-box transcription factor T | | Authors: | Bebenek, A, Linhares, B, Jaime-Figueroa, S, Butrin, A, Crews, C. | | Deposit date: | 2022-12-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of a Small Molecule Downmodulator for the Transcription Factor Brachyury.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6QNZ
 
 | | Crystal structure of the site-specific DNA nickase N.BspD6I E418A Mutant | | Descriptor: | GLYCEROL, Heterodimeric restriction endonuclease R.BspD6I large subunit, PHOSPHATE ION | | Authors: | Artyukh, R.I, Kachalova, G.S, Yunusova, A.K, Gabdulkhakov, A.G, Fatkhullin, B.F, Atanasov, B.P, Perevyazova, T.A, Popov, A.N, Zheleznaya, L.A. | | Deposit date: | 2019-02-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The key role of E418 carboxyl group in the formation of Nt.BspD6I nickase active site: Structural and functional properties of Nt.BspD6I E418A mutant.
J.Struct.Biol., 210, 2020
|
|
6TEP
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6SJ6
 
 | | Cryo-EM structure of 50S-RsfS complex from Staphylococcus aureus | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Khusainov, I, Pellegrino, S, Yusupova, G, Yusupov, M, Fatkhullin, B. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
1RJD
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity
J.Biol.Chem., 279, 2004
|
|
5LUC
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at 1.8 Angstrom; internal aldimine with PLP in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Cellini, B, Borri Voltattorni, C, Montioli, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
1RJE
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity.
J.Biol.Chem., 279, 2004
|
|
6H7W
 
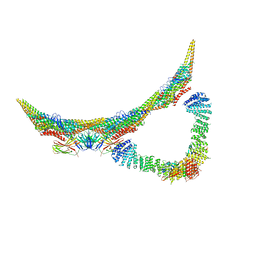 | | Model of retromer-Vps5 complex assembled on membrane. | | Descriptor: | Putative vacuolar protein sorting-associated protein, Vacuolar protein sorting-associated protein 26-like protein, Vacuolar protein sorting-associated protein 29, ... | | Authors: | Kovtun, O, Leneva, N, Ariotti, N, Rohan, T.S, Owen, D.J, Briggs, J.A.G, Collins, B.M. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
4YDU
 
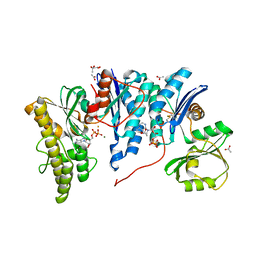 | | Crystal structure of E. coli YgjD-YeaZ heterodimer in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W, Collinet, B, Perrochia, L, Durand, D, Van Tilbeurgh, H. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5AH8
 
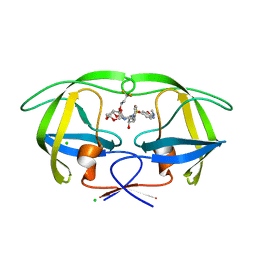 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(3,3,3-trifluoropropoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AHA
 
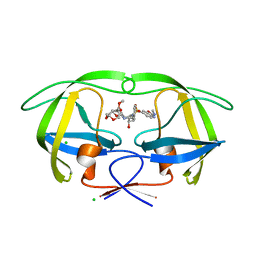 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH7
 
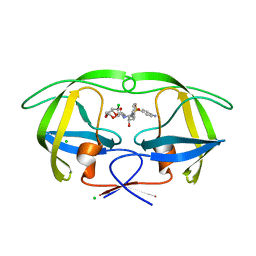 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH6
 
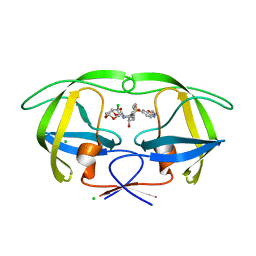 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5AHC
 
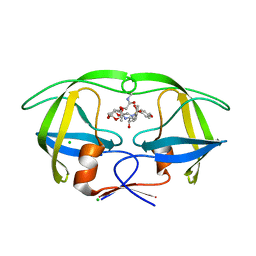 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
1R9M
 
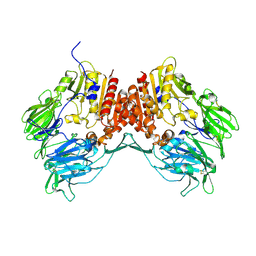 | | Crystal Structure of Human Dipeptidyl Peptidase IV at 2.1 Ang. Resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, ... | | Authors: | Aertgeerts, K, Ye, S, Tennant, M.G, Collins, B, Rogers, J, Sang, B.C, Skene, R.J, Webb, D.R, Prasad, G.S. | | Deposit date: | 2003-10-30 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation.
Protein Sci., 13, 2004
|
|
5AHB
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[(2Z)-2-(methylimino)-2,3-dihydro-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AGZ
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[(2-fluoroprop-2-en-1-yl)oxy]hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[2-(methylamino)-1,3-benzoxazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-04 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH9
 
 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-(2-methoxyethoxy)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Enstrom, O, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
