3X06
 
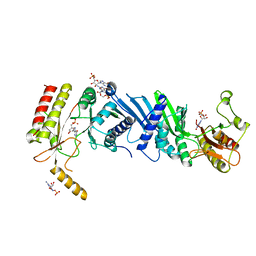 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X05
 
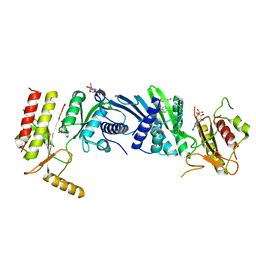 | | Crystal structure of PIP4KIIBETA T201M complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X09
 
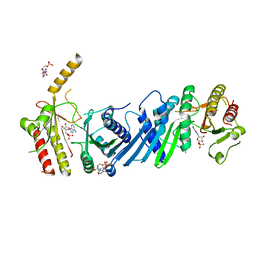 | | Crystal structure of PIP4KIIBETA F205L complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X08
 
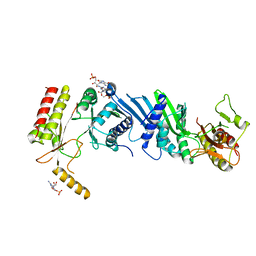 | | Crystal structure of PIP4KIIBETA N203A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0B
 
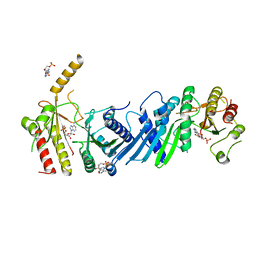 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0C
 
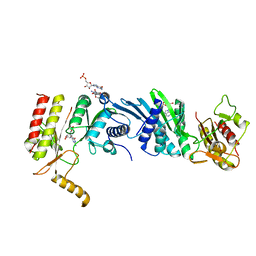 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X07
 
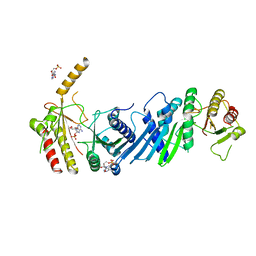 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
6MBI
 
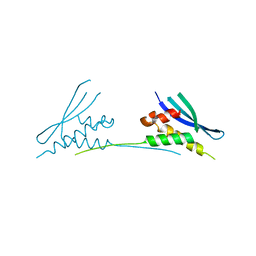 | |
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | Descriptor: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2AYL
 
 | | 2.0 Angstrom Crystal Structure of Manganese Protoporphyrin IX-reconstituted Ovine Prostaglandin H2 Synthase-1 Complexed With Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, GLYCEROL, ... | | Authors: | Gupta, K, Selinsky, B.S, Loll, P.J. | | Deposit date: | 2005-09-07 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7P4R
 
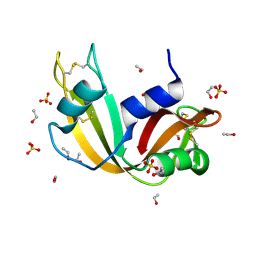 | | Ultra High Resolution X-ray Structure of Orthorhombic Bovine Pancreatic Ribonuclease at 100K | | Descriptor: | ETHANOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Lisgarten, D.R, Palmer, R.A, Cooper, J.B, Naylor, C.E, Howlin, B.J, Lisgarten, J.N, Najmudin, S, Lobley, C.M.C. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultra-high resolution X-ray structure of orthorhombic bovine pancreatic Ribonuclease A at 100K.
BMC Chem, 17, 2023
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
5UF1
 
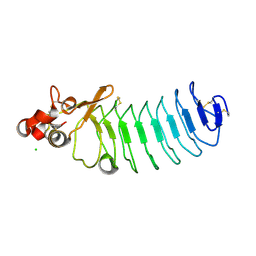 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) O13 in complex with H-trisaccharide | | Descriptor: | CHLORIDE ION, O13, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gunn, R.J, Collins, B.C, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
6XS7
 
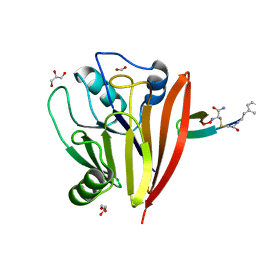 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-D2 | | Descriptor: | 48V-DTY-THR-THR-ILE-TYR-TRP-THR-PRO-LEU-GLY-THR-PHE-PRO-ARG-ILE-ARG, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XSA
 
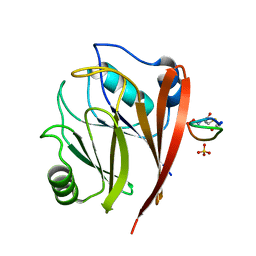 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L2 | | Descriptor: | 48V-TYR-LEU-PRO-THR-ILE-THR-GLY-VAL-GLY-HIS-LEU-TRP-HIS-PRO-LEU, SULFATE ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XS5
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-D1 | | Descriptor: | 48V-DTY-ILE-ILE-ASP-THR-PRO-LEU-GLY-VAL-PHE-LEU-SER-SER-LEU-LYS-ARG, FORMIC ACID, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XS9
 
 | | Crystal structure of human Vps29 complexed with RaPID-derived cyclic peptide RT-L1 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 48V-TYR-ILE-LYS-THR-PRO-LEU-GLY-THR-PHE-PRO-ASN-ARG-HIS-GLY, GLYCEROL, ... | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
6XS8
 
 | | Crystal structure of Chaetomium thermophilum Vps29 complexed with RaPID-derived cyclic peptide RT-D3 | | Descriptor: | 48V-DTY-GLY-TYR-ASP-PRO-LEU-GLY-LEU-LYS-TYR-PHE-ALA, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, K.-E, Guo, Q, Collins, B.M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95009851 Å) | | Cite: | De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex.
Sci Adv, 7, 2021
|
|
1XE7
 
 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANINE, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
3HMJ
 
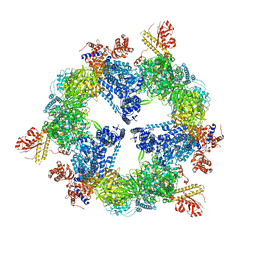 | | Saccharomyces cerevisiae FAS type I | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, P, Mulinacci, B, Koestler, C, Vollrath, R, Oesterhelt, D, Grininger, M. | | Deposit date: | 2009-05-29 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Structure, 17, 2009
|
|
8ESD
 
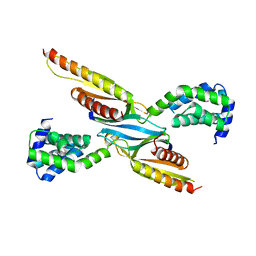 | | Crystal structure of COMMD7-COMMD9-COMMD5-COMMD10 tetramer | | Descriptor: | COMM domain-containing protein 10, COMM domain-containing protein 5, COMM domain-containing protein 7, ... | | Authors: | Healy, M.D, Collins, B.M. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8ESE
 
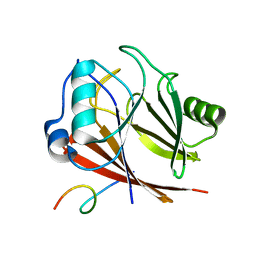 | |
8F2R
 
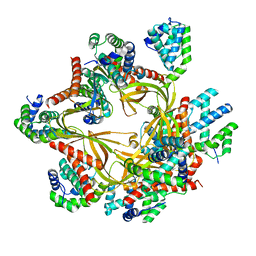 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8F2U
 
 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
