7WJH
 
 | | Crystal structure of Bcl-xL bound to the BH3 domain of human Pxt1 | | Descriptor: | Bcl-2-like protein 1, MAGNESIUM ION, Peroxisomal testis-specific protein 1, ... | | Authors: | Lim, D, Ku, B. | | Deposit date: | 2022-01-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structural and biochemical analyses of Bcl-xL in complex with the BH3 domain of peroxisomal testis-specific 1.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
8GSV
 
 | | Crystal structure of human BAK in complex with the Pxt1 BH3 domain | | Descriptor: | Bcl-2 homologous antagonist/killer, Peroxisomal testis-specific protein 1 | | Authors: | Lim, D, Ku, B. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for proapoptotic activation of Bak by the noncanonical BH3-only protein Pxt1.
Plos Biol., 21, 2023
|
|
2HB5
 
 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
4JRN
 
 | | ROP18 kinase domain in complex with AMP-PNP and sucrose | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Rhoptry kinase family protein, ... | | Authors: | Lim, D, Gold, D.A, Lindsay, J, Rosowski, E.E, Niedelman, W, Yaffe, M.B, Saeij, J.P.J. | | Deposit date: | 2013-03-21 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the Toxoplasma gondii ROP18 kinase domain reveals a second ligand binding pocket required for acute virulence.
J.Biol.Chem., 288, 2013
|
|
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKL
 
 | |
1DKO
 
 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKP
 
 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKN
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1VQQ
 
 | |
1G68
 
 | | PSE-4 CARBENICILLINASE, WILD TYPE | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
1G6A
 
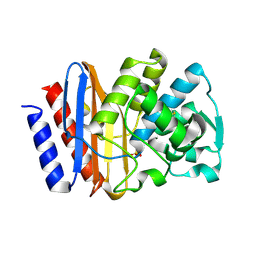 | | PSE-4 CARBENICILLINASE, R234K MUTANT | | Descriptor: | BETA-LACTAMASE PSE-4, SULFATE ION | | Authors: | Lim, D, Sanschagrin, F, Passmore, L, De Castro, L, Levesque, R.C, Strynadka, N.C.J. | | Deposit date: | 2000-11-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies.
Biochemistry, 40, 2001
|
|
1K3E
 
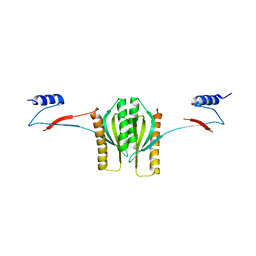 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3S
 
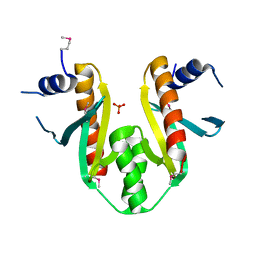 | | Type III Secretion Chaperone SigE | | Descriptor: | PHOSPHATE ION, SigE | | Authors: | Bertero, M.G, Luo, Y, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-28 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
2OLU
 
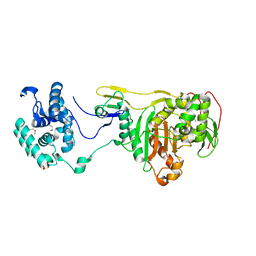 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L.H, Lim, D, Strynadka, N.C. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2OLV
 
 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L, Lim, D, Strynadka, N.C.J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
8IGC
 
 | | Crystal structure of Bak bound to Bnip5 BH3 | | Descriptor: | Bcl-2 homologous antagonist/killer, Protein BNIP5 | | Authors: | Ku, B, Lim, D. | | Deposit date: | 2023-02-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Crystal structure of Bak bound to the BH3 domain of Bnip5, a noncanonical BH3 domain-containing protein.
Proteins, 92, 2024
|
|
7D35
 
 | | Human LC8 bound to ebola virus VP35(67-76) | | Descriptor: | Dynein light chain 1, cytoplasmic, Peptide from Polymerase cofactor VP35 | | Authors: | Ku, B, Lim, D. | | Deposit date: | 2020-09-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of human LC8 bound to a peptide from Ebola virus VP35.
J.Microbiol, 59, 2021
|
|
6JM4
 
 | |
4DFW
 
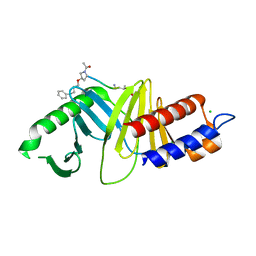 | | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides | | Descriptor: | CHLORIDE ION, Peptide, Serine/threonine-protein kinase PLK1 | | Authors: | Liu, F, Park, J.-E, Qian, W.-J, Lim, D, Gr ber, M, Berg, T, Yaffe, M.B, Lee, K.S, Burke Jr, T.R. | | Deposit date: | 2012-01-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Oxime-based Post Solid-phase Peptide Diversification: Identification of High Affinity Polo-like Kinase 1 (Plk1) Polo-box Domain Binding Peptides
Chem.Biol., 2012
|
|
4JIZ
 
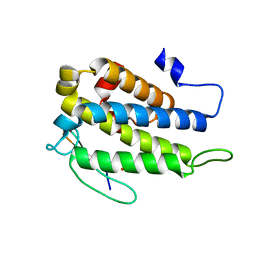 | | Human Mob1-phosphopeptide complex | | Descriptor: | MOB kinase activator 1A, ZINC ION, phosphopeptide | | Authors: | Stach, L, Ogrodowicz, R.W, Rock, J.M, Lim, D, Yaffe, M.B, Amon, A, Smerdon, S.J. | | Deposit date: | 2013-03-07 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes.
Science, 340, 2013
|
|
1MDW
 
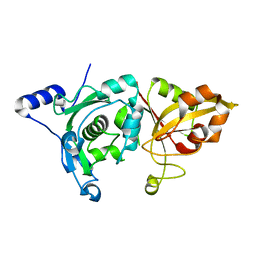 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
1KXR
 
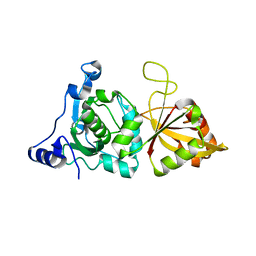 | | Crystal Structure of Calcium-Bound Protease Core of Calpain I | | Descriptor: | CALCIUM ION, thiol protease DOMAINS I AND II | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Elce, J.S, Jia, Z, Davies, P.L. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Ca(2+) switch aligns the active site of calpain.
Cell(Cambridge,Mass.), 108, 2002
|
|
1RO8
 
 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
