6GRC
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Sodium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRD
 
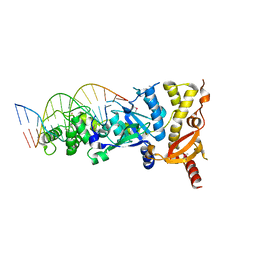 | | eukaryotic junction-resolving enzyme GEN-1 binding with Cesium | | Descriptor: | CESIUM ION, DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRB
 
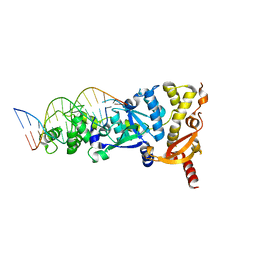 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | Descriptor: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
5T5A
 
 | |
6R47
 
 | |
4CS1
 
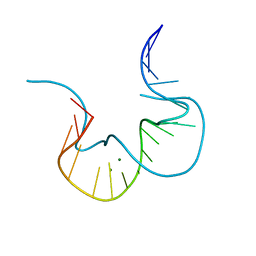 | |
4OJI
 
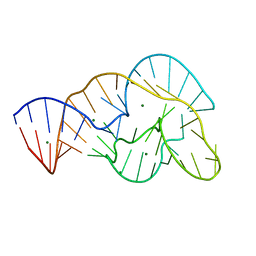 | | Crystal Structure of Twister Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (52-MER) | | Authors: | Liu, Y, Wilson, T.J, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2014-01-21 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and mechanistic investigation of the twister ribozyme.
Nat.Chem.Biol., 10, 2014
|
|
8ITS
 
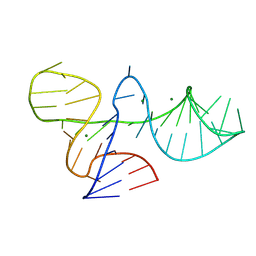 | |
7V9E
 
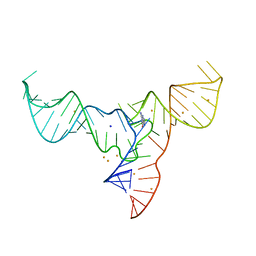 | | Crystal structure of a methyl transferase ribozyme | | Descriptor: | BARIUM ION, GUANINE, RNA (68-MER), ... | | Authors: | Deng, J, Lilley, D.M.J, Huang, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a methyltransferase ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
5CNQ
 
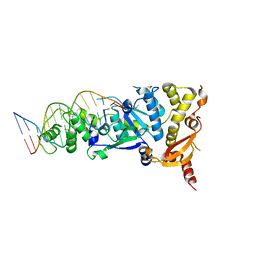 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5CO8
 
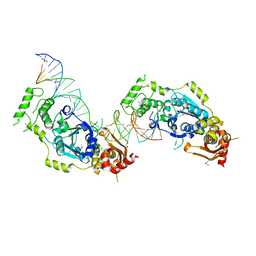 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
352D
 
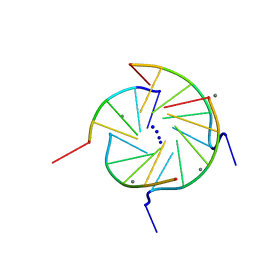 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
6TB7
 
 | |
6TF2
 
 | |
6TFH
 
 | |
8HBA
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HB8
 
 | |
8HB3
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
6Q8U
 
 | |
6Q8V
 
 | |
8I3Z
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN at 1.67 angstrom | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3'), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
7EAG
 
 | | Crystal structure of the RAGATH-18 k-turn | | Descriptor: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
2YDH
 
 | |
