5WAX
 
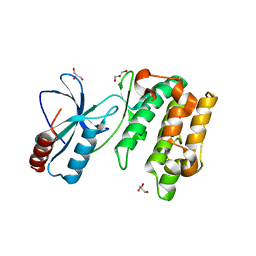 | | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10) | | Descriptor: | GLYCEROL, SAPK10 (serine/threonine-protein kinase 10) | | Authors: | Righetto, G.L, Counago, R.M, Halabelian, L, Santiago, A.S, Massirer, K.B, Arruda, P, Gileadi, O, Menossi, M, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10)
To Be Published
|
|
7MEX
 
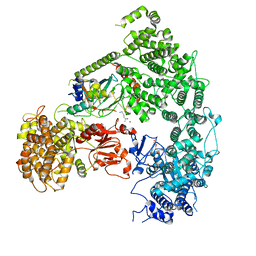 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
1VYH
 
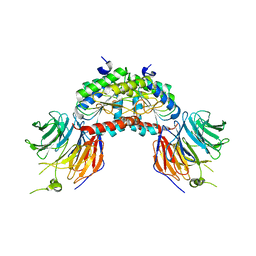 | | PAF-AH Holoenzyme: Lis1/Alfa2 | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB ALPHA SUBUNIT, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB BETA SUBUNIT | | Authors: | Tarricone, C, Perrina, F, Monzani, S, Massimiliano, L, Knapp, S, Tsai, L.-H, Derewenda, Z.S, Musacchio, A. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Coupling Paf Signaling to Dynein Regulation: Structure of Lis1 in Complex with Paf-Acetylhydrolase.
Neuron, 44, 2004
|
|
2JQG
 
 | | Leader Protease | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
4J59
 
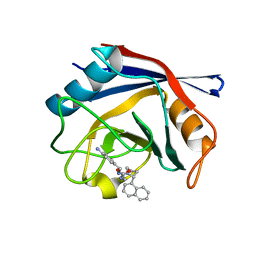 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(naphthalen-1-yl)pyrrolidin-1-yl]-2-oxoethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
4J5A
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(2,5-dimethoxyphenyl)pyrrolidin-1-yl]-2-oxoethyl}urea, DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Rationnal Design of small-molecule inhibitors of human Cyclophilins and HCV replication.
to be published
|
|
2JQF
 
 | | Full Length Leader Protease of Foot and Mouth Disease Virus C51A Mutant | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
1UNL
 
 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | Descriptor: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
5VGT
 
 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1UNG
 
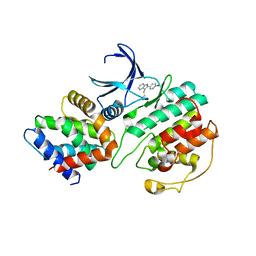 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
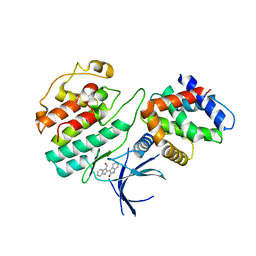 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
7M3X
 
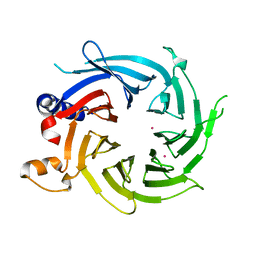 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
4J5C
 
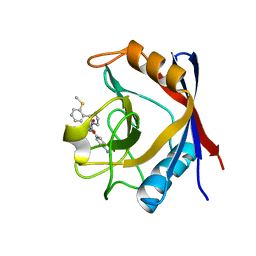 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-[(2S)-4-(methylsulfanyl)-1-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-1-oxobutan-2-yl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
4J5E
 
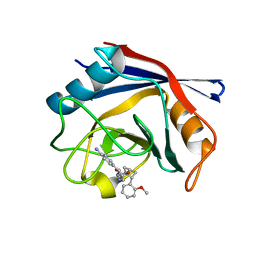 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(2-methoxyphenyl)pyrrolidin-1-yl]-2-oxoethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1EF7
 
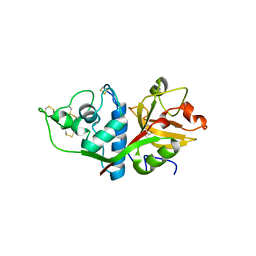 | | CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X | | Descriptor: | CATHEPSIN X | | Authors: | Guncar, G, Klemencic, I, Turk, B, Turk, V, Karaoglanovic-Carmona, A, Juliano, L, Turk, D. | | Deposit date: | 2000-02-07 | | Release date: | 2000-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease.
Structure Fold.Des., 8, 2000
|
|
7U9I
 
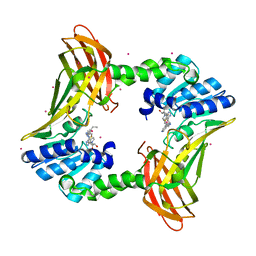 | | Co-crystal structure of human CARM1 in complex with MT556 inhibitor | | Descriptor: | 7-[5-S-(4-{[(4-ethylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-arginine methyltransferase CARM1, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Perveen, S, Dong, A, Hutchinson, A, Seitova, A, Gibson, E, Hajian, T, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure of human CARM1 in complex with MT556 inhibitor
To Be Published
|
|
2MI7
 
 | | Solution NMR structure of alpha3Y | | Descriptor: | de novo protein a3Y | | Authors: | Glover, S.D, Jorge, C, Liang, L, Valentine, K.G, Hammarstrom, L, Tommos, C. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photochemical tyrosine oxidation in the structurally well-defined alpha 3Y protein: proton-coupled electron transfer and a long-lived tyrosine radical.
J.Am.Chem.Soc., 136, 2014
|
|
2AA1
 
 | | Crystal structure of the cathodic hemoglobin isolated from the Antarctic fish Trematomus Newnesi | | Descriptor: | Hemoglobin alpha-1 chain, Hemoglobin beta-C chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Bonomi, G, Lubrano, M.C, Merlino, A, Riccio, A, Vergara, A, Vitagliano, L, Verde, C, Di Prisco, G. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Minimal structural requirements for root effect: crystal structure of the cathodic hemoglobin isolated from the antarctic fish Trematomus newnesi
Proteins, 62, 2006
|
|
1R5D
 
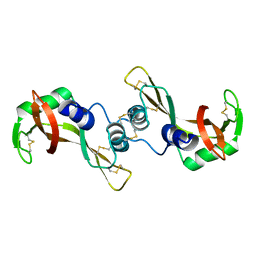 | | X-ray structure of bovine seminal ribonuclease swapping dimer from a new crystal form | | Descriptor: | Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
1R5C
 
 | | X-ray structure of the complex of Bovine seminal ribonuclease swapping dimer with d(CpA) | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
3TCO
 
 | | Crystallographic and spectroscopic characterization of Sulfolobus solfataricus TrxA1 provide insights into the determinants of thioredoxin fold stability | | Descriptor: | 1,2-ETHANEDIOL, Thioredoxin (TrxA-1) | | Authors: | Esposito, L, Ruggiero, A, Masullo, M, Ruocco, M.R, Lamberti, A, Arcari, P, Zagari, A, Vitagliano, L. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and spectroscopic characterizations of Sulfolobus solfataricus TrxA1 provide insights into the determinants of thioredoxin fold stability.
J.Struct.Biol., 177, 2012
|
|
2LXY
 
 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
7UW7
 
 | | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of E3 ubiquitin-protein ligase TRIP12, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Dong, A, Li, Y, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP
To Be Published
|
|
7MEQ
 
 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
4QBB
 
 | | Structure of the foot-and-mouth disease virus leader proteinase in complex with inhibitor (N~2~-[(3S)-4-({(2R)-1-[(4-CARBAMIMIDAMIDOBUTYL)AMINO]-4-METHYL-1-OXOPENTAN-2-YL}AMINO)-3-HYDROXY-4-OXOBUTANOYL]-L-ARGINYL-L-PROLINAMIDE) | | Descriptor: | Leader protease, N~2~-[(3S)-4-({(2R)-1-[(4-carbamimidamidobutyl)amino]-4-methyl-1-oxopentan-2-yl}amino)-3-hydroxy-4-oxobutanoyl]-L-arginyl-L-prolinamide, PHOSPHATE ION, ... | | Authors: | Grishkovskaya, I, Steinberger, J, Cencic, R, Juliano, M.A, Juliano, L, Skern, T. | | Deposit date: | 2014-05-07 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Foot-and-mouth disease virus leader proteinase: Structural insights into the mechanism of intermolecular cleavage.
Virology, 468-470C, 2014
|
|
