7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC4
 
 | | Crystal structure of serotonin 2A receptor in complex with serotonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
8WWS
 
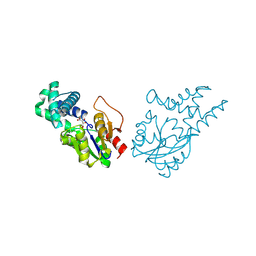 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
8XU8
 
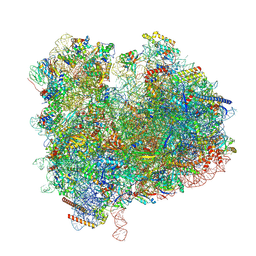 | | State 2c(S2c) of yeast 80S ribosome bound to compact eEF2 and 2 tRNAs during peptidyl transferation | | Descriptor: | 18S rRNA, 25S rRNA, 5.8S rRNA, ... | | Authors: | Cheng, J, Wu, C.L, Li, J.X, Zhang, X.Z. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Snapshots of ribosome dynamics near atomic resolution in situ: a full insight into eukaryotic elongation cycle
To Be Published
|
|
8YLR
 
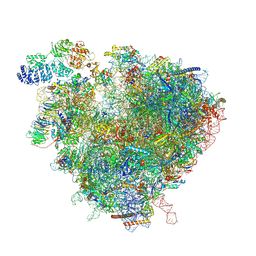 | | State 6 (S6) of yeast 80S ribosome bound to 2 tRNAs and eEF2 and eEF3 during tranlocation | | Descriptor: | 18S rRNA, 25S rRNA, 5.8S rRNA, ... | | Authors: | Cheng, J, Wu, C.L, Li, J.X, Zhang, X.Z. | | Deposit date: | 2024-03-06 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Snapshots of ribosome dynamics near atomic resolution in situ: a full insight into eukaryotic elongation cycle
To Be Published
|
|
8Z70
 
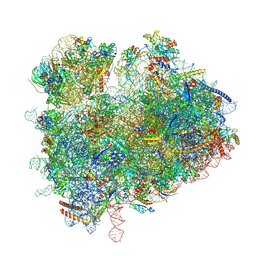 | | State 1 (S1) of yeast 80S ribosome bound to 2 tRNAs during mRNA decoding | | Descriptor: | 18S rRNA, 25S rRNA, 5.8S rRNA, ... | | Authors: | Cheng, J, Wu, C.L, Li, J.X, Zhang, X.Z. | | Deposit date: | 2024-04-19 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Snapshots of ribosome dynamics near atomic resolution in situ: a full insight into eukaryotic elongation cycle
To Be Published
|
|
8Z71
 
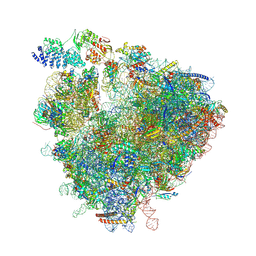 | | State 1a (S1a) of yeast 80S ribosome bound to open eEF3 and 2 tRNAs and eEF1A during mRNA decoding | | Descriptor: | 18S rRNA, 25S rRNA, 5.8S rRNA, ... | | Authors: | Cheng, J, Wu, C.L, Li, J.X, Zhang, X.Z. | | Deposit date: | 2024-04-19 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Snapshots of ribosome dynamics near atomic resolution in situ: a full insight into eukaryotic elongation cycle
To Be Published
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8W1W
 
 | |
7DR8
 
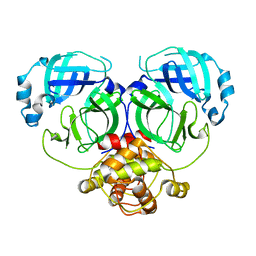 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-26 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.338149 Å) | | Cite: | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
7DR9
 
 | |
8W1X
 
 | |
8WF7
 
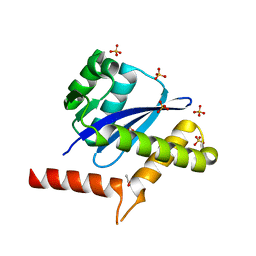 | | The Crystal Structure of integrase from Biortus | | Descriptor: | ACETATE ION, Integrase, SULFATE ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of integrase from Biortus
To Be Published
|
|
8WF4
 
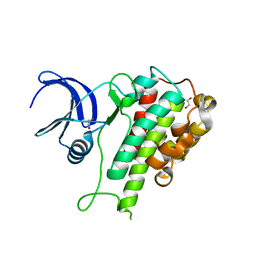 | | The Crystal Structure of RSK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Ribosomal protein S6 kinase alpha-1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of RSK1 from Biortus.
To Be Published
|
|
8WFY
 
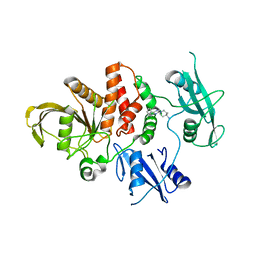 | | The Crystal Structure of SHP2 from Biortus. | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of SHP2 from Biortus.
To Be Published
|
|
8X5M
 
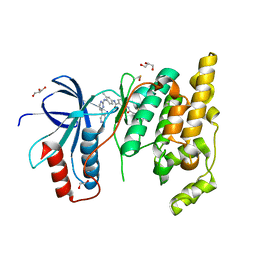 | | The Crystal Structure of JNK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(dimethylamino)butanoylamino]-~{N}-[3-methyl-4-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]benzamide, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of JNK1 from Biortus.
To Be Published
|
|
7VBD
 
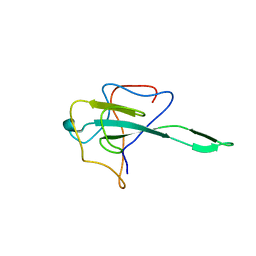 | | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0 | | Descriptor: | Nucleoprotein | | Authors: | Zeng, P, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0
To Be Published
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7VBF
 
 | |
7VBE
 
 | |
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F4G
 
 | | Structure of RPAP2-bound RNA polymerase II | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-06-18 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
7FJ2
 
 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
