5X3V
 
 | | Structure of human SHMT2 protein mutant | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase, mitochondrial | | Authors: | Li, J, Sheng, J. | | Deposit date: | 2017-02-09 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of human SHMT2 protein mutant
To Be Published
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
4JN9
 
 | | Crystal structure of the DepH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
4JNA
 
 | | Crystal structure of the DepH complex with dimethyl-FK228 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
4L93
 
 | | Crystal structure of Human Hsp90 with S36 | | Descriptor: | 3,4-dihydroisoquinolin-2(1H)-yl[2,4-dihydroxy-5-(propan-2-yl)phenyl]methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Crystal structure of Human Hsp90 with S36
To be Published
|
|
8UOZ
 
 | | EmrE structure in the TPP-bound state (WT/E14Q heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE, TETRAPHENYLPHOSPHONIUM | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
5DTH
 
 | |
8UWU
 
 | | EmrE structure in the proton-bound state (WT/L51I heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
4L90
 
 | | Crystal structure of Human Hsp90 with RL3 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Human Hsp90 with RL3
to be published
|
|
8HHE
 
 | |
5XBF
 
 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XSJ
 
 | | XylFII-LytSN complex | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS, ... | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IDC
 
 | |
8IGQ
 
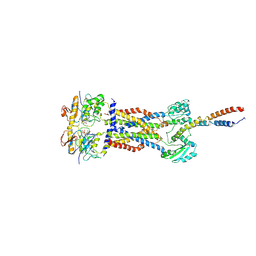 | | Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
8IDD
 
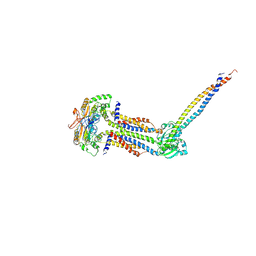 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
8IDB
 
 | |
8JIA
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, Luo, M. | | Deposit date: | 2023-05-26 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis.
Nat Commun, 14, 2023
|
|
8XAS
 
 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5ZCK
 
 | | Structure of the RIP3 core region | | Descriptor: | SODIUM ION, peptide from Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Li, J, Wu, H. | | Deposit date: | 2018-02-18 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex.
Cell, 173, 2018
|
|
5WSV
 
 | | Crystal structure of Myosin VIIa IQ5 in complex with Ca2+-CaM | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
8XAT
 
 | |
7CFU
 
 | |
8X5N
 
 | | Tetramer Gabija with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | tetramer Gabija with ATP (local refinement) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
