6W62
 
 | | Cryo-EM structure of Cas12i-crRNA complex | | Descriptor: | Cas12i, crRNA | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.916 Å) | | Cite: | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
6W5C
 
 | | Cryo-EM structure of Cas12i(E894A)-crRNA-dsDNA complex | | Descriptor: | Cas12i, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6W64
 
 | |
5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A6G
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, S1-S4 DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
7JZY
 
 | | CryoEM structure of a CRISPR-Cas complex | | Descriptor: | AcrF9, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of a CRISPR-Cas complex
To Be Published
|
|
7L48
 
 | | Cryo-EM structure of a CRISPR-Cas12f Binary Complex | | Descriptor: | Cas12f, ZINC ION, sgRNA | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
7L49
 
 | | Cryo-EM structure of CRISPR-Cas12f Ternary Complex | | Descriptor: | Cas12f1, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
7JZX
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF7 | | Descriptor: | AcrF7, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZW
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF4 | | Descriptor: | CRISPR repeat sequence, CRISPR type I-F/YPEST-associated protein Csy1, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A22
 
 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
2N96
 
 | | An unexpected mode of small molecule DNA binding provides the structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A | | Descriptor: | (1R,1'R,2S,2'S,3R,3'R,5aR,10aR,11a'S)-2'-[(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)oxy]-2,2'-diethyl-11,11 '-dihydrazinyl-6,6',9,9'-tetrahydroxy-4,4',5,5',10,10'-hexaoxo-1,1'-bis{[2,4,6-trideoxy-4-(dimethylamino)-beta-L-arabino -hexopyranosyl]oxy}[2,2',3,3',4,4',5,5',5a,8,10,10',10a,11a'-tetradecahydro-1H,1'H-[3,3'-bibenzo[b]fluorene]]-2-yl 2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranoside, DNA (5'-D(*GP*CP*TP*AP*TP*AP*GP*C)-3') | | Authors: | Woo, C.M, Li, Z, Paulson, E, Herzon, S.B. | | Deposit date: | 2015-11-07 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5A6E
 
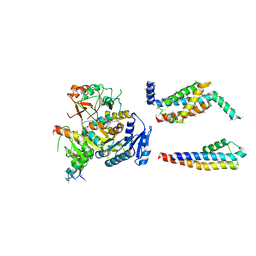 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
7XT3
 
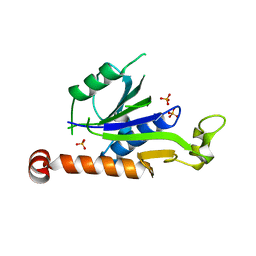 | | Crystal Structure of Hepatitis virus A 2C protein 128-335 aa | | Descriptor: | Genome polyprotein, PHOSPHATE ION | | Authors: | Chen, P, Wojdyla, J.A, Li, Z, Wang, M, Cui, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural characterization of hepatitis A virus 2C reveals an unusual ribonuclease activity on single-stranded RNA.
Nucleic Acids Res., 50, 2022
|
|
7XVE
 
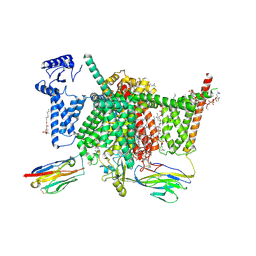 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
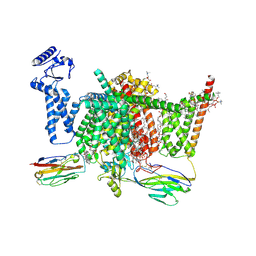 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8IN8
 
 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I88
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
6DU8
 
 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
