8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14182 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8I41
 
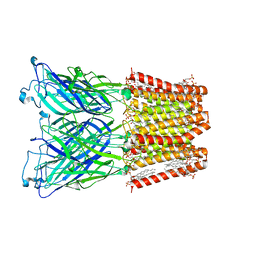 | |
8I42
 
 | |
8I48
 
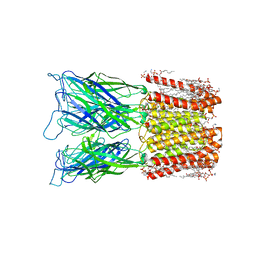 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-01-18 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8I47
 
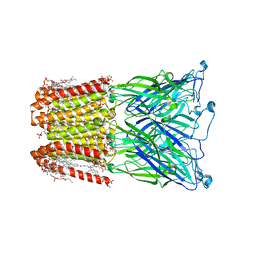 | |
7WDL
 
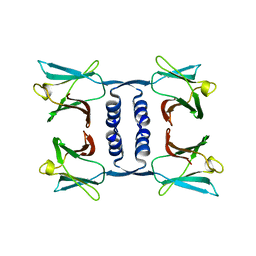 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
7UVB
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT021601 | | Descriptor: | 2-hydroxy-6-({(3S)-4-[2-(2-hydroxyethyl)pyridine-3-carbonyl]morpholin-3-yl}methoxy)benzaldehyde, FORMYL GROUP, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Kaya, E, Xu, Q, Li, Z, Strutt, S.C, Cathers, B.E. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GBT021601 improves red blood cell health and the pathophysiology of sickle cell disease in a murine model.
Br.J.Haematol., 202, 2023
|
|
7VTV
 
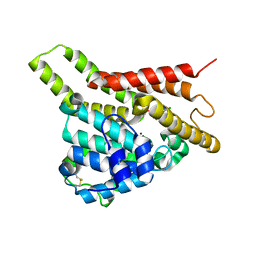 | | Crystal structure of PDE8A catalytic domain in complex with 15 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-(difluoromethoxy)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VTX
 
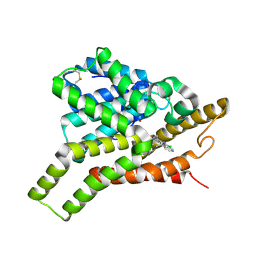 | | Crystal structure of PDE8A catalytic domain in complex with 22 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-(pyridin-4-yl)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.50010943 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VSL
 
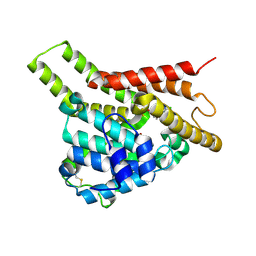 | | Crystal structure of PDE8A catalytic domain in complex with 10 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)benzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.500069 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7VTW
 
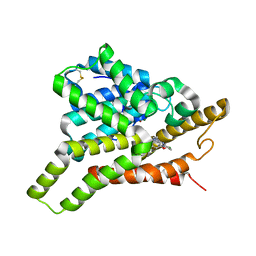 | | Crystal structure of PDE8A catalytic domain in complex with 17 | | Descriptor: | 2-chloro-9-(3-(2,2-difluoroethoxy)-5-isopropoxybenzyl)-9H-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wu, X.-N, Zhou, Q, Huang, Y.-D, Li, Z, Wu, Y, Luo, H.-B. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79971337 Å) | | Cite: | Structure-Based Discovery of Orally Efficient PDE8 Inhibitors for the Treatment of Vascular Dementia
To Be Published
|
|
7DTC
 
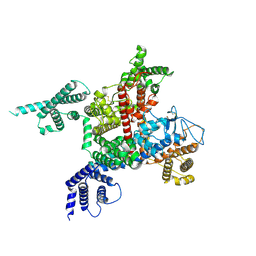 | | voltage-gated sodium channel Nav1.5-E1784K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Pan, X, Li, Z. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na v 1.5 reveals the fast inactivation-related segments as a mutational hotspot for the long QT syndrome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7F0D
 
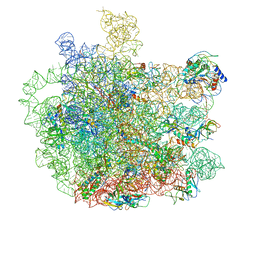 | | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosome subunit bound with clarithromycin | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Zhang, W, Sun, Y, Gao, N, Li, Z. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides.
Emerg Microbes Infect, 11, 2022
|
|
7CWH
 
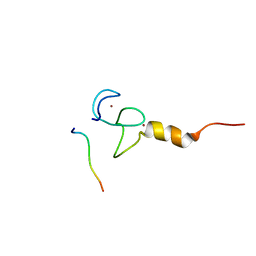 | | Structural basis of RACK7 PHD to read a pediatric glioblastoma-associated histone mutation H3.3G34R | | Descriptor: | Peptide from Histone H3.3, Protein kinase C-binding protein 1, ZINC ION | | Authors: | Lan, W.X, Li, Z, Jiao, F.F, Wang, C.X, Guo, R, Cao, C.Y. | | Deposit date: | 2020-08-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of RACK7 PHD domain to read a pediatric glioblastoma‐associated histone mutation H3.3G34R
Chin.J.Chem., 2021
|
|
1ULW
 
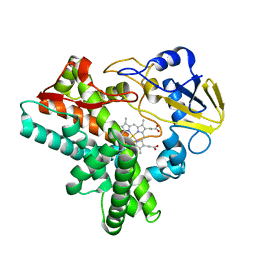 | | Crystal structure of P450nor Ser73Gly/Ser75Gly mutant | | Descriptor: | Cytochrome P450 55A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Oshima, R, Fushinobu, S, Su, F, Li, Z, Takaya, N, Shoun, H. | | Deposit date: | 2003-09-16 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for direct hydride transfer from NADH to cytochrome P450nor
J.Mol.Biol., 342, 2004
|
|
5X3P
 
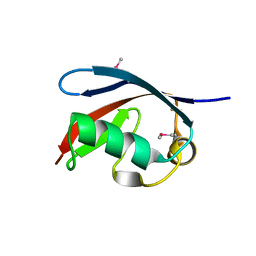 | |
5X4L
 
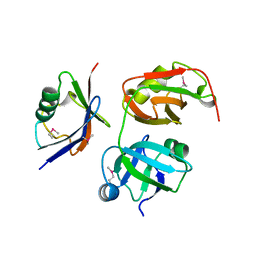 | | Crystal structure of the UBX domain of human UBXD7 in complex with p97 N domain | | Descriptor: | Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 7 | | Authors: | Jiang, T, Li, Z, Wang, Y, Xu, M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structures of the UBX domain of human UBXD7 and its complex with p97 ATPase
Biochem. Biophys. Res. Commun., 486, 2017
|
|
7W1I
 
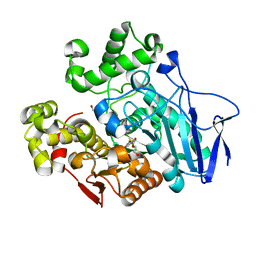 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1J
 
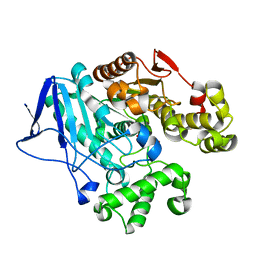 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1L
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X | | Descriptor: | Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
