7D8L
 
 | |
7D8I
 
 | |
7D8Q
 
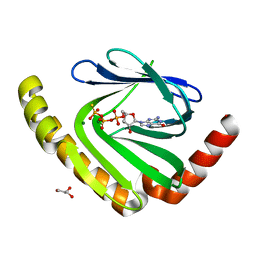
 | | The structure of nucleotide phosphatase Sa1684 complex with GDP analogue from Staphylococcus aureus | | Descriptor: | MAGNESIUM ION, UPF0374 protein SAB1800c, [(2R,3R,4S,5S)-5-(2-azanyl-6-oxidanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl bis(oxidanyl)phosphinothioyl hydrogen phosphate | | Authors: | Wang, Z, Li, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural mechanism for the nucleoside tri- and diphosphate hydrolysis activity of Ntdp from Staphylococcus aureus.
Febs J., 288, 2021
|
|
7FHO
 
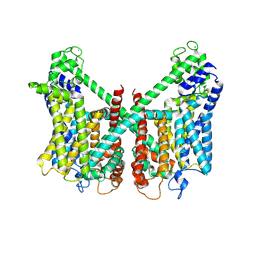 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 50 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHN
 
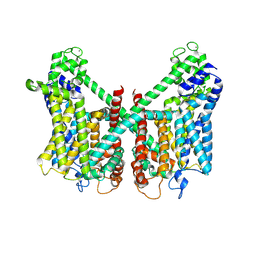 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHL
 
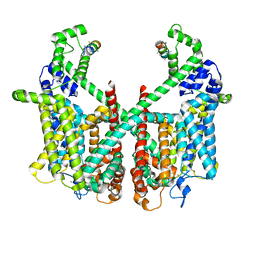 | | Structure of AtTPC1 with 50 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHK
 
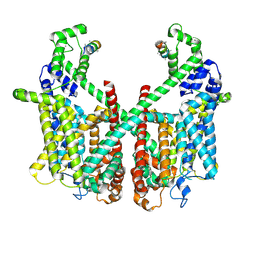 | | Structure of AtTPC1 with 1 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2GVE
 
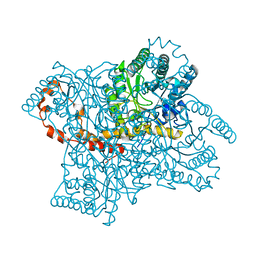 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GO4
 
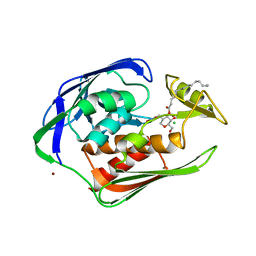 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
2K3C
 
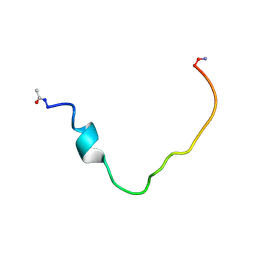 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
7F0X
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F12
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F15
 
 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
2KBV
 
 | | Structural and functional analysis of TM XI of the NHE1 isoform of thE NA+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Lee, B.L, Li, X, Liu, Y, Sykes, B.D, Fliegel, L. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane XI of the NHE1 Isoform of the Na+/H+ Exchanger
J.Biol.Chem., 284, 2009
|
|
7XIO
 
 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
5CXV
 
 | | Structure of the human M1 muscarinic acetylcholine receptor bound to antagonist Tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, 1,2-ETHANEDIOL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, B, Feng, D, Li, X, Kobilka, T.S, Kobilka, B.K. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-09 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the M1 and M4 muscarinic acetylcholine receptors.
Nature, 531, 2016
|
|
8KCZ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
8JU4
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
