2HNU
 
 | | Crystal Structure of a Dipeptide Complex of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNV
 
 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNW
 
 | | Crystal Structure of the F91STOP mutant of des1-6 Bovine Neurophysin-I, unliganded state | | Descriptor: | Oxytocin-neurophysin 1 | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
8IBI
 
 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IBJ
 
 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IAN
 
 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
7EQH
 
 | |
7VBQ
 
 | | Heterodimer structure of Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxIJ | | Descriptor: | FE (III) ION, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxI, Fe(II)/(alpha)ketoglutarate-dependent dioxygenase TlxJ, ... | | Authors: | Li, X, Awakawa, T, Mori, T, Abe, I. | | Deposit date: | 2021-09-01 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heterodimeric Non-heme Iron Enzymes in Fungal Meroterpenoid Biosynthesis.
J.Am.Chem.Soc., 143, 2021
|
|
7VBR
 
 | |
7WJG
 
 | | Streptococcus mutans BusR transcriptional factor | | Descriptor: | CITRIC ACID, Putative transcriptional regulator (GntR family) | | Authors: | Li, X, Liu, N. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural study of transcriptional factor BusR
To Be Published
|
|
7XIN
 
 | | Crystal structure of DODC from Pseudomonas | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Li, X, Zhou, Y.L, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Sun, D.Y, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DODC from Pseudomonas
To Be Published
|
|
7CR2
 
 | | human KCNQ2 in complex with retigabine | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR7
 
 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR0
 
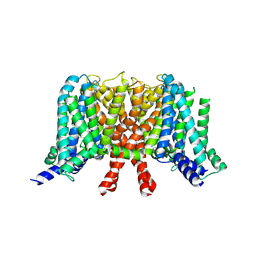 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
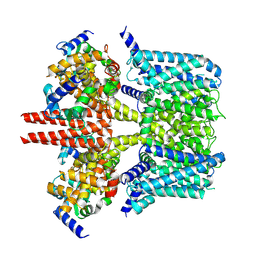 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR4
 
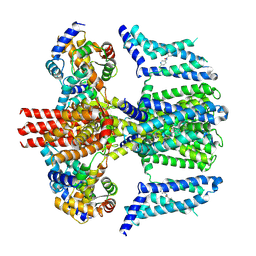 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
8JQE
 
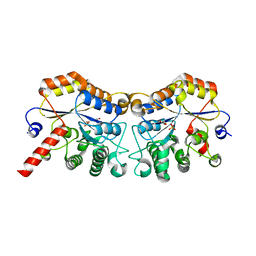 | | Structure of CmCBDA in complex with Mn2+ and glycerol | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
8JQF
 
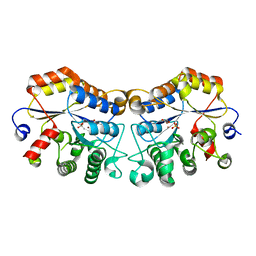 | | Structure of CmCBDA in complex with Ni2+ and Glycerol | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
7VS7
 
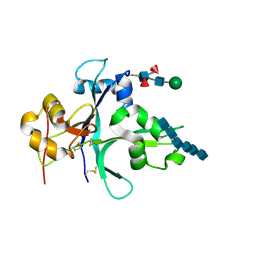 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
1VM2
 
 | |
1VM4
 
 | |
4NXL
 
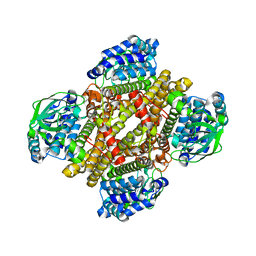 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
