7XEE
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with 2-(3-phenyloxetan-3-yl)ethanamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-phenyloxetan-3-yl)ethanamine, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XEF
 
 | | Crystal Structure of the Y53F/N55A mutant of LEH complexed with (R)-(1-benzyl-3-phenylpyrrolidin-3-yl)methanol | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Limonene-1,2-epoxide hydrolase, ... | | Authors: | Qu, G, Li, X, Sun, Z.T. | | Deposit date: | 2022-03-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | Authors: | Yan, X, Li, X, Song, G. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
2OBR
 
 | | Crystal Structures of P Domain of Norovirus VA387 | | Descriptor: | Capsid protein | | Authors: | Cao, S, Lou, Z, Jiang, X, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2006-12-20 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of blood group trisaccharides by norovirus.
J.Virol., 81, 2007
|
|
7YPG
 
 | |
7F7G
 
 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
2R5W
 
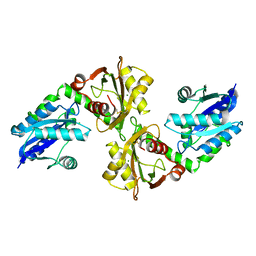 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
7CLA
 
 | |
8JBZ
 
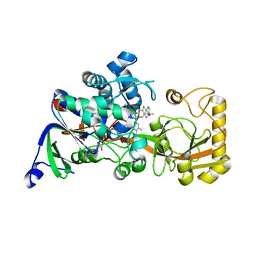 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, 4-ANDROSTENE-3-17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 4-androstadiene-3,17- dione
To Be Published
|
|
8JOJ
 
 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate | | Descriptor: | 3-ketosteroid dehydrogenase, 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.819 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 9,11-epoxy-17-hydroxypregn-4-ene-3,20-dione actate
To Be Published
|
|
8JTR
 
 | | Cryo-EM structure of GeoCas9-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA (139-bp) | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-22 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of GeoCas9-sgRNA binary complex
To Be Published
|
|
8JTJ
 
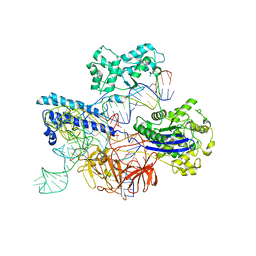 | | Cryo-EM structure of GeoCas9-sgRNA-dsDNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (29-MER), DNA (5'-D(P*GP*GP*GP*CP*GP*CP*GP*AP*A)-3'), ... | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure of GeoCas9-sgRNA-DNA ternary complex
To Be Published
|
|
2OBT
 
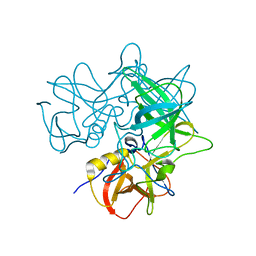 | |
7EXT
 
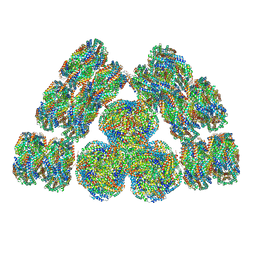 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
7EYD
 
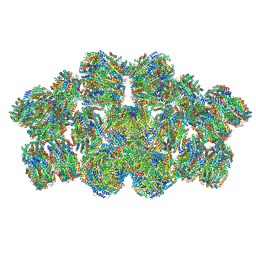 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | Descriptor: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-30 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
4IF2
 
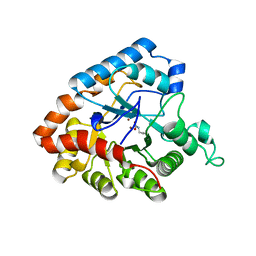 | |
2OBS
 
 | |
8YJA
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain lacking beta-flap | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MARTX cysteine protease domain, SODIUM ION | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
8Z0L
 
 | | Cryo-EM structure of Cas8-HNH system at partial R-loop state | | Descriptor: | DNA (32-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8Z0K
 
 | | Cryo-EM structure of Cas8-HNH system at full R-loop state | | Descriptor: | DNA (37-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
8ZDY
 
 | | Cryo-EM structure of Cas8-HNH system at target free state | | Descriptor: | RNA (58-MER), a protein | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8ZNR
 
 | | Cryo-EM structure of Cas8-HNH system at ssDNA-bound state | | Descriptor: | DNA (35-MER), RNA (56-MER), protein structure | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
7W7F
 
 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
