4CO3
 
 | |
4CO0
 
 | |
6OEU
 
 | | Structure of human Patched1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Qi, X, Li, X, Wang, J. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6KJ2
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from single crystal at 0.67 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.67 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
8W51
 
 | |
4CO1
 
 | |
4CNY
 
 | |
4CO2
 
 | |
6KJ1
 
 | | 200kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.65 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.65 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6KJ3
 
 | | 120kV MicroED structure of FUS (37-42) SYSGYS solved from merged datasets at 0.60 A | | Descriptor: | RNA-binding protein FUS | | Authors: | Zhou, H, Luo, F, Luo, Z, Li, D, Liu, C, Li, X. | | Deposit date: | 2019-07-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.6 Å) | | Cite: | Programming Conventional Electron Microscopes for Solving Ultrahigh-Resolution Structures of Small and Macro-Molecules.
Anal.Chem., 91, 2019
|
|
4M58
 
 | | Crystal Structure of an transition metal transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | Authors: | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
4CO5
 
 | |
4DM4
 
 | | The conserved domain of yeast Cdc73 | | Descriptor: | Cell division control protein 73 | | Authors: | Chen, H, Shi, N, Gao, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3S5J
 
 | |
6M1Y
 
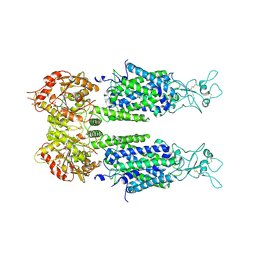 | | The overall structure of KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
8KGN
 
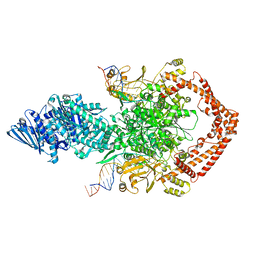 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGL
 
 | |
8KGO
 
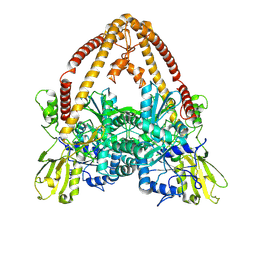 | |
8KGR
 
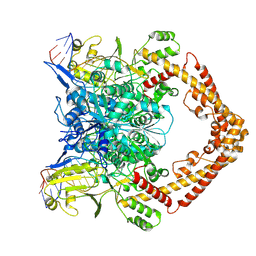 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (32-MER), DNA (33-MER), DNA topoisomerase 2, ... | | Authors: | Cong, J, Xin, U, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGM
 
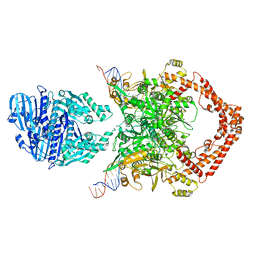 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGP
 
 | |
