7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
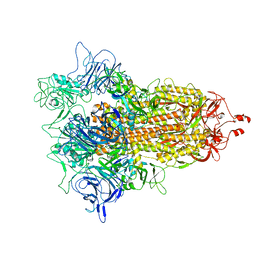 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | Descriptor: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7FJ3
 
 | | Cryo-EM structure of PRV A-capid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
1ROW
 
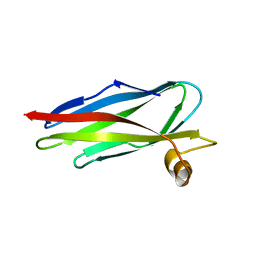 | | Structure of SSP-19, an MSP-domain protein like family member in Caenorhabditis elegans | | Descriptor: | MSP-domain protein like family member | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, J, Lu, S, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7UU4
 
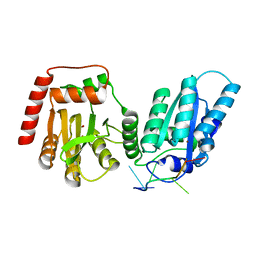 | | Crystal structure of APOBEC3G complex with ssRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU3
 
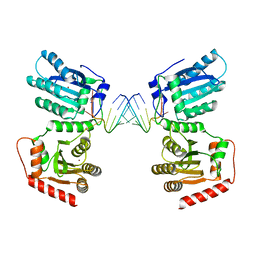 | | Crystal structure of APOBEC3G complex with 3'overhangs RNA-Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(*CP*CP*CP*AP*CP*GP*GP*GP*AP*AP*U)-3'), RNA (5'-R(*CP*CP*CP*GP*UP*GP*GP*GP*AP*AP*U)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU5
 
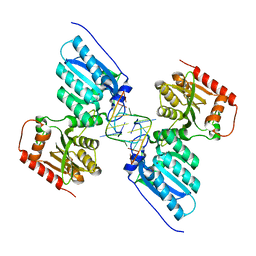 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
6JIP
 
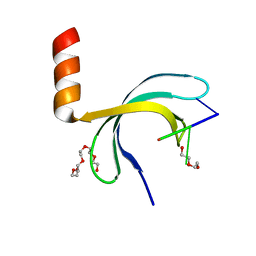 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIQ
 
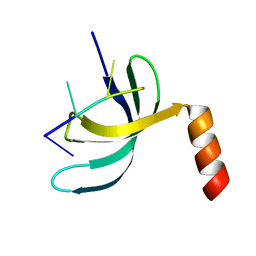 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZHZ
 
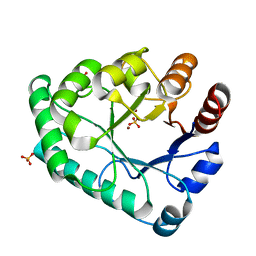 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKM
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
7CJQ
 
 | | Structure of DLA-88*001:04 | | Descriptor: | ARG-THR-ILE-SER-TYR-THR-TYR-PRO-PHE, Beta-2-microglobulin, MHC class I DLA-88 | | Authors: | Sun, Y.J, Ma, L.Z, Li, S. | | Deposit date: | 2020-07-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of canine MHC class I at 2.7 Angstroms resolution
To Be Published
|
|
7CFD
 
 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | Descriptor: | FI20010p1, Protein aubergine | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFB
 
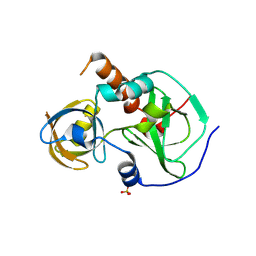 | | Drosophila melanogaster Krimper eTud1 apo structure | | Descriptor: | FI20010p1, SULFATE ION | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
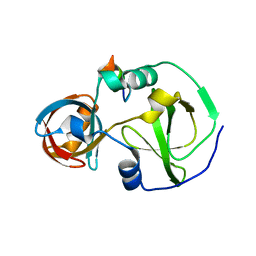 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | Descriptor: | FI20010p1, Protein argonaute-3 | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7CZB
 
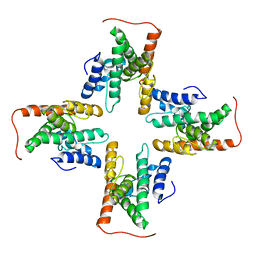 | | The cryo-EM structure of the ERAD retrotranslocation channel formed by human Derlin-1 | | Descriptor: | Derlin-1 | | Authors: | Rao, B, Li, S, Yao, D, Wang, Q, Xia, Y, Jia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2020-09-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The cryo-EM structure of an ERAD protein channel formed by tetrameric human Derlin-1.
Sci Adv, 7, 2021
|
|
7FBI
 
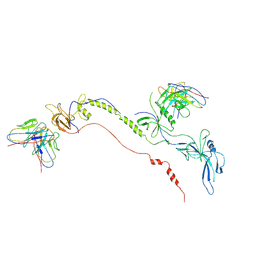 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
7F3X
 
 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7EWT
 
 | | The crystal structure of Lysophospholipid acyltransferase LPCAT3 (MOBAT5) in its monomeric and apo form | | Descriptor: | Lysophospholipid acyltransferase 5 | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
