7XBN
 
 | | Crystal Structure of YC-17-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
1FNJ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
8ZXH
 
 | |
8XFF
 
 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein in complex with SPR phage tail tube protein | | Descriptor: | DSR2(H171A), SPR tail tube protein | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
3KS4
 
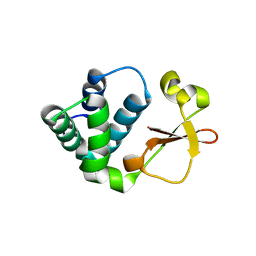 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
7WLU
 
 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.81 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
5TGZ
 
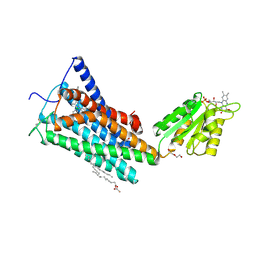 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
3KS8
 
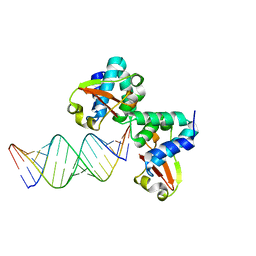 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
2C6H
 
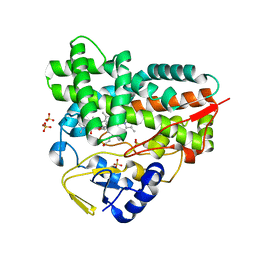 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
4G8O
 
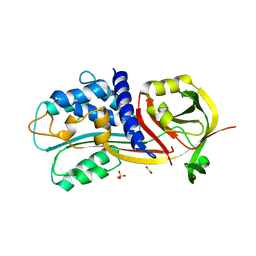 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), 1,2-ETHANEDIOL, Plasminogen activator inhibitor 1, ... | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2KWN
 
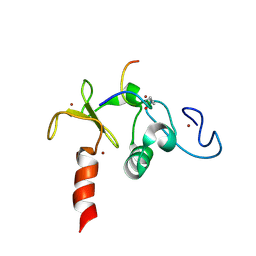 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing acetylation at Lysine 16 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWO
 
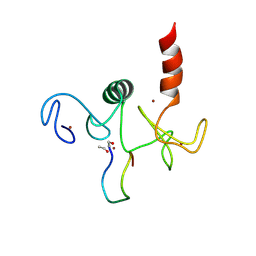 | | Solution structure of the double PHD (plant homeodomain) fingers of human transcriptional protein DPF3b bound to a histone H4 peptide containing N-terminal acetylation at Serine 1 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-14 | | Release date: | 2010-07-14 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KWK
 
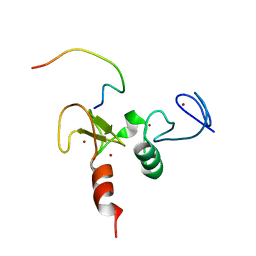 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3b bound to a H3 peptide wild type | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
6NYG
 
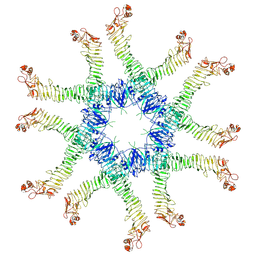 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2a (OA-2a) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8YG8
 
 | | The early intermediate structure of baculovirus fusion protein GP64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major envelope glycoprotein | | Authors: | Du, D, Guo, J, Li, S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch.
Nat Commun, 15, 2024
|
|
8YG6
 
 | | The pre-fusion structure of baculovirus fusion protein GP64 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Major envelope glycoprotein | | Authors: | Du, D, Guo, J, Li, S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural transition of GP64 triggered by a pH-sensitive multi-histidine switch.
Nat Commun, 15, 2024
|
|
6NYJ
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
