7WSF
 
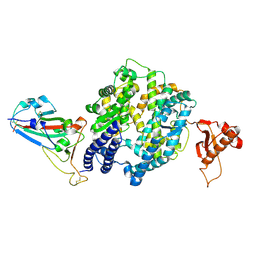 | |
3SQ9
 
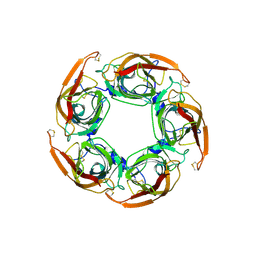 | | Crystal Structures of the Ligand Binding Domain of a Pentameric Alpha7 Nicotinic Receptor Chimera | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neuronal acetylcholine receptor subunit alpha-7, ... | | Authors: | Li, S.-X, Huang, S, Bren, N, Noridomi, K, Dellisanti, C, Sine, S, Chen, L. | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ligand-binding domain of an alpha 7-nicotinic receptor chimera and its complex with agonist.
Nat.Neurosci., 14, 2011
|
|
3SQ6
 
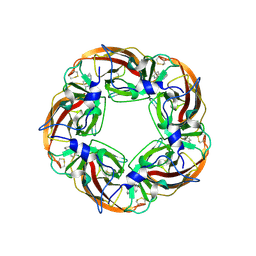 | | Crystal Structures of the Ligand Binding Domain of a Pentameric Alpha7 Nicotinic Receptor Chimera with its Agonist Epibatidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIBATIDINE, ... | | Authors: | Li, S.-X, Huang, S, Bren, N, Noridomi, K, Dellisanti, C, Sine, S, Chen, L. | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand-binding domain of an alpha 7-nicotinic receptor chimera and its complex with agonist.
Nat.Neurosci., 14, 2011
|
|
7YGB
 
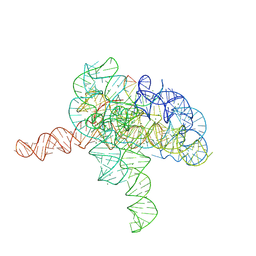 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
4HQP
 
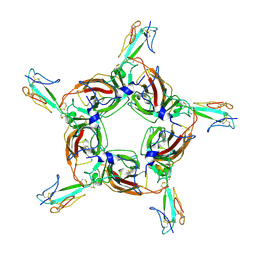 | | Alpha7 nicotinic receptor chimera and its complex with Alpha bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Li, S.X, Cheng, K, Gomoto, R, Bren, N, Huang, S, Sine, S, Chen, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural principles for Alpha-neurotoxin binding to and selectivity among nicotinic receptors
To be Published
|
|
7WSH
 
 | | Cryo-EM structure of SARS-CoV-2 spike receptor-binding domain in complex with sea lion ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Li, S, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cross-species recognition and molecular basis of SARS-CoV-2 and SARS-CoV binding to ACE2s of marine animals.
Natl Sci Rev, 9, 2022
|
|
7WSE
 
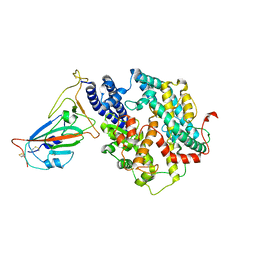 | |
7EHL
 
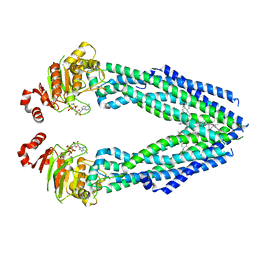 | | Cryo-EM structure of human ABCB8 transporter in nucleotide binding state | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, Mitochondrial potassium channel ATP-binding subunit, ... | | Authors: | Li, S.J, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY | | Cite: | Cryo-EM structure of human ABCB8 transporter in nucleotide binding state.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
8IZ1
 
 | |
8IYY
 
 | |
8IZ0
 
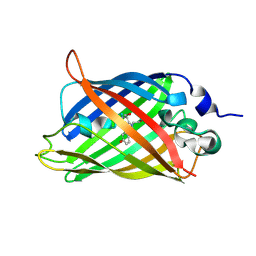 | | mTurquoise2 W66Y | | Descriptor: | Green fluorescent protein | | Authors: | Li, S.A, Kang, J.S. | | Deposit date: | 2023-04-06 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unified intracellular pH spectrum with sitepHorin-a quantum-entanglement-based pH-sensitive and ratiometric fluorescent protein
To Be Published
|
|
8K3Y
 
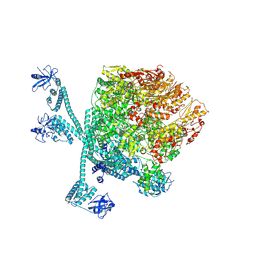 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
7CQI
 
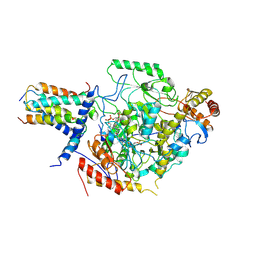 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CQK
 
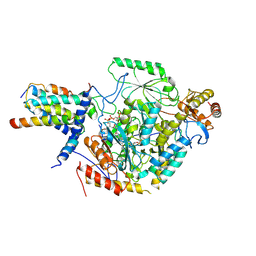 | | Cryo-EM structure of the substrate-bound SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, Serine palmitoyltransferase 1, Serine palmitoyltransferase 2, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-08-11 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7FD4
 
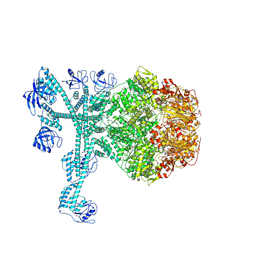 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FID
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-08-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
