1EGU
 
 | | CRYSTAL STRUCTURE OF STREPTOCOCCUS PNEUMONIAE HYALURONATE LYASE AT 1.56 A RESOLUTION | | Descriptor: | HYALURONATE LYASE, SULFATE ION | | Authors: | Li, S, Kelly, S.J, Lamani, E, Ferraroni, M, Jedrzejas, M.J. | | Deposit date: | 2000-02-16 | | Release date: | 2001-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase.
EMBO J., 19, 2000
|
|
2Y5Z
 
 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y46
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in C 2 2 21 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y5N
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
5Y9F
 
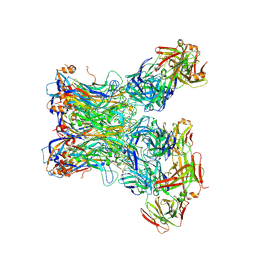 | | Crystal structure of HPV59 pentamer in complex with the Fab fragment of antibody 28F10 | | Descriptor: | Major capsid protein L1, heavy chain of Fab fragment of antibody 28F10, light chains of Fab fragment of antibody 28F10 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
5GVU
 
 | |
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9KPN
 
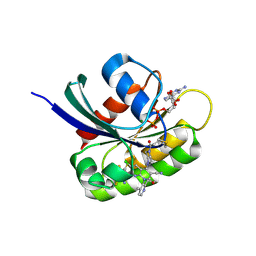 | | Crystal structure of KRAS-G12C in complex with Compound 20 (JAB-20) | | Descriptor: | 7-[2-azanyl-3,4,5,6-tetrakis(fluoranyl)phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | Deposit date: | 2024-11-23 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
9KPM
 
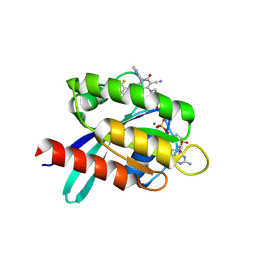 | | Crystal structure of KRAS-G12C in complex with compound 16 (JAB-16) | | Descriptor: | 7-[2-azanyl-3,5-bis(chloranyl)-6-fluoranyl-phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | Deposit date: | 2024-11-23 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
7PYV
 
 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
1F1S
 
 | |
7YG8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
2VZM
 
 | |
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
2VZ7
 
 | | Crystal structure of the YC-17-bound PikC D50N mutant | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, S, Sherman, D.H, Podust, L.M. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Analysis of Transient and Catalytic Desosamine Binding Pockets in Cytochrome P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 284, 2009
|
|
4ZHW
 
 | |
9LHJ
 
 | | UBE2N/UBE2V2 complexed with a covalent inhibitor | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Li, S, Wu, X, Zhou, L, Lu, X. | | Deposit date: | 2025-01-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Proteome-Wide Data Guides the Discovery of Lysine-Targeting Covalent Inhibitors Using DNA-Encoded Chemical Libraries.
Angew.Chem.Int.Ed.Engl., 2025
|
|
9LG2
 
 | |
1F9G
 
 | |
5F22
 
 | | C-terminal domain of SARS-CoV nsp8 complex with nsp7 | | Descriptor: | Non-structural protein, Non-structural protein 7 | | Authors: | Li, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | C-terminal domain of SARS-CoV nsp8 complex with nsp7
To Be Published
|
|
