5Z3O
 
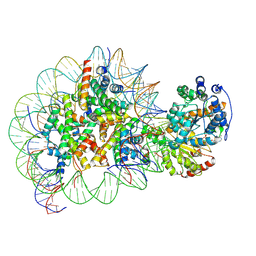 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
1DPJ
 
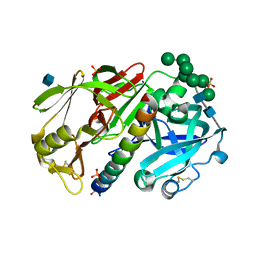 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
8IP4
 
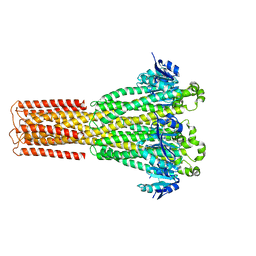 | | Cryo-EM structure of hMRS-highEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP5
 
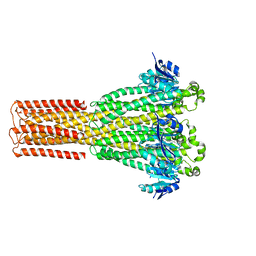 | | Cryo-EM structure of hMRS2-lowEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP6
 
 | | Cryo-EM structure of hMRS2-rest | | Descriptor: | CHLORIDE ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP3
 
 | | Cryo-EM structure of hMRS2-Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
5Z3L
 
 | | Structure of Snf2-nucleosome complex in apo state | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
2QOP
 
 | | Crystal structure of the transcriptional regulator AcrR from Escherichia coli | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Li, M, Gu, R, Su, C.-C, McDermott, G, Yu, E.W. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the transcriptional regulator AcrR from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
5C9B
 
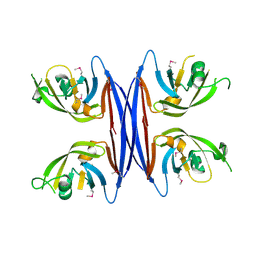 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5C9D
 
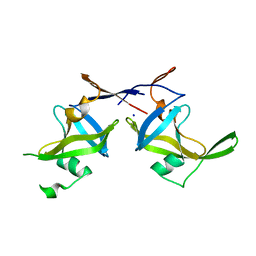 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6IY3
 
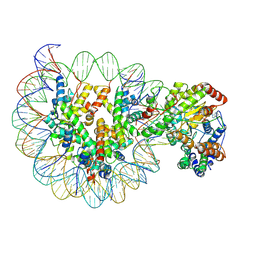 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
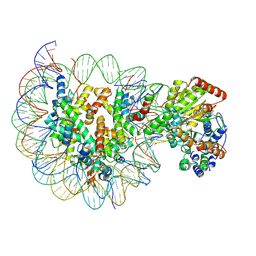 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5C9F
 
 | | Crystal structure of a retropepsin-like aspartic protease from Rickettsia conorii | | Descriptor: | ApRick protease, CHLORIDE ION, SODIUM ION | | Authors: | Li, M, Gustchina, A, Cruz, R, Simoes, M, Curto, P, Martinez, J, Faro, C, Simoes, I, Wlodawer, A. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of RC1339/APRc from Rickettsia conorii, a retropepsin-like aspartic protease.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6VDK
 
 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
3LIZ
 
 | | crystal structure of bla g 2 complexed with Fab 4C3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C3 monoclonal antibody Heavy Chain, ... | | Authors: | Li, M, Gustchina, A, Glesner, J, Wunschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of allergen-antibody interaction of cockroach allergen Bla g 2 with monoclonal antibodies that inhibit IgE antibody binding.
Plos One, 6, 2011
|
|
7JOD
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein 4 (Prostase, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JOW
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein 4 (Prostase, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JQO
 
 | | Crystal structure of the R64D mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JRX
 
 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides complexed with Bovine Chymotrypsin | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JOS
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | Kallikrein 4 (Prostase, enamel matrix, prostate), ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-07 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JOE
 
 | | Crystal structure of BbKI complexed with Human Kallikrein 4 | | Descriptor: | Kallikrein 4 (Prostase, enamel matrix, prostate), ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JQN
 
 | | Crystal structure of the R64M mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JR2
 
 | |
7JR1
 
 | | Crystal structure of the R64F mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Bovine Trypsin | | Descriptor: | Cationic trypsin, Kunitz-type inihibitor, SODIUM ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7JQK
 
 | | Crystal structure of the R64A mutant of Bauhinia Bauhinioides Kallikrein Inhibitor complexed with Human Kallikrein 4 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Kallikrein-4, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A. | | Deposit date: | 2020-08-11 | | Release date: | 2021-07-21 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural studies of complexes of kallikrein 4 with wild-type and mutated forms of the Kunitz-type inhibitor BbKI.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
