8FNL
 
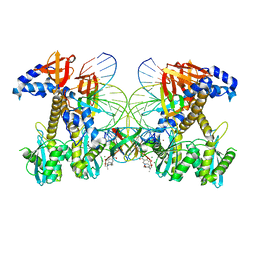 | | Structure of E138K/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNG
 
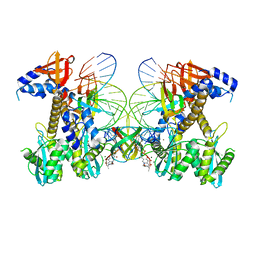 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNM
 
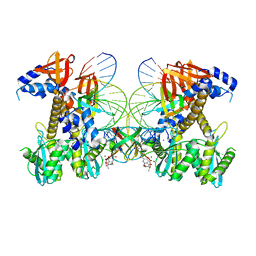 | | Structure of G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNQ
 
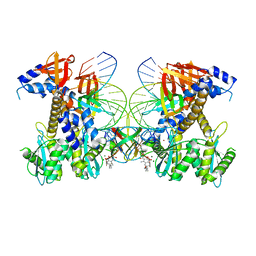 | | Structure of E138K/G140A/Q148K HIV-1 intasome with 4d bound | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FND
 
 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNO
 
 | | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNH
 
 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
4RC5
 
 | |
4RI2
 
 | | Crystal structure of the photoprotective protein PsbS from spinach | | Descriptor: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RI3
 
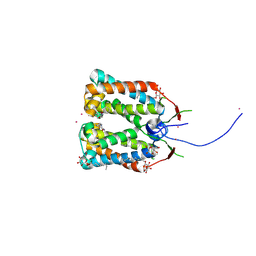 | | Crystal structure of DCCD-modified PsbS from spinach | | Descriptor: | DICYCLOHEXYLUREA, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6E4H
 
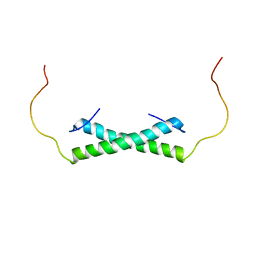 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
6X5N
 
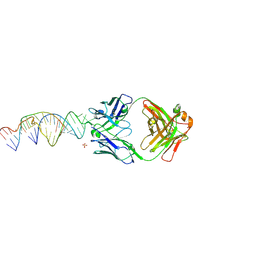 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6X5M
 
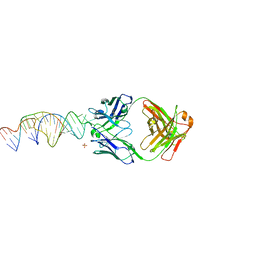 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
4Z5S
 
 | |
5O9I
 
 | | Crystal structure of transcription factor IIB Mvu mini-intein | | Descriptor: | Transcription initiation factor IIB,Transcription initiation factor IIB | | Authors: | Mikula, K.M, Iwai, H, Li, M, Wlodawer, A. | | Deposit date: | 2017-06-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
6KY4
 
 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
1Z28
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1Z29
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ACETIC ACID, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
7F79
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F77
 
 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | Descriptor: | Glutamate dehydrogenase | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
2A3R
 
 | | Crystal Structure of Human Sulfotransferase SULT1A3 in Complex with Dopamine and 3-Phosphoadenosine 5-Phosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, L-DOPAMINE, Monoamine-sulfating phenol sulfotransferase | | Authors: | Lu, J.H, Li, H.T, Liu, M.C, Zhang, J.P, Li, M, An, X.M, Chang, W.R. | | Deposit date: | 2005-06-26 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3'-phosphoadenosine 5'-phosphate
Biochem.Biophys.Res.Commun., 335, 2005
|
|
1FMX
 
 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1FMU
 
 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
1GA1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF IODOTYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FRAGMENT OF IODOTYROSTATIN, ... | | Authors: | Dauter, Z, Li, M, Wlodawer, A. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
