6LOX
 
 | | Crystal Structure of human glutaminase with macrocyclic inhibitor | | Descriptor: | (E)-15,22-Dioxa-4,11-diaza-5(2,5)-thiadiazola-10(3,6)-pyridazina-1,14(1,3)-dibenzenacyclodocosaphan-18-ene-3,12-dione, Glutaminase kidney isoform, mitochondrial | | Authors: | Bian, J, Li, Z, Xu, X, Wang, J, Li, L. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Enabled Discovery of Novel Macrocyclic Inhibitors Targeting Glutaminase 1 Allosteric Binding Site.
J.Med.Chem., 64, 2021
|
|
3HBF
 
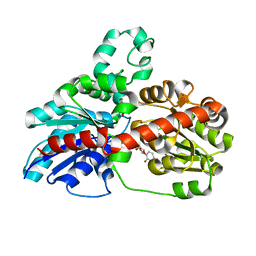 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
3HBJ
 
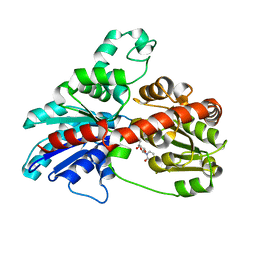 | | Structure of UGT78G1 complexed with UDP | | Descriptor: | Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
4IVT
 
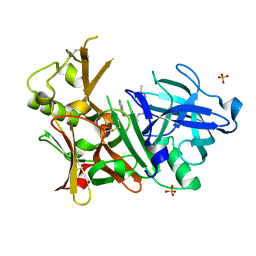 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(1H-indol-1-yl)acetamide, SULFATE ION | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
4IVS
 
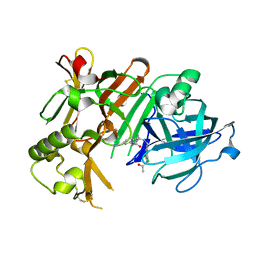 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-{N-[4-(acetylamino)-3,5-dichlorobenzyl]carbamimidoyl}-2-(6-cyano-1H-indol-1-yl)acetamide | | Authors: | Chen, T.T, Li, L, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2013-01-23 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors
Molecules, 18, 2013
|
|
8IWD
 
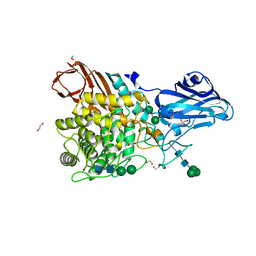 | | Aspergillus niger Rha-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
8IWF
 
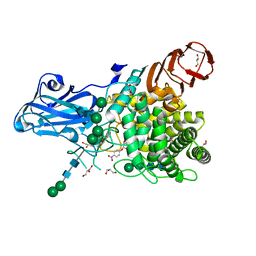 | | Aspergillus niger Rha-2 and pNPR | | Descriptor: | (2S,3R,4R,5R,6S)-2-methyl-6-[4-[oxidanyl(oxidanylidene)-$l^4-azanyl]phenoxy]oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, L.J, Li, L, Wang, M.H, Jiang, Z.D, Zhu, Y.B, Jin, T.C, Ni, H. | | Deposit date: | 2023-03-29 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure and catalytic function of alpha-L-rhamnosidase from Aspergillus niger
To Be Published
|
|
4FQB
 
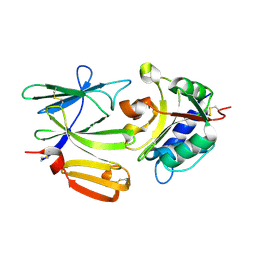 | |
4FCO
 
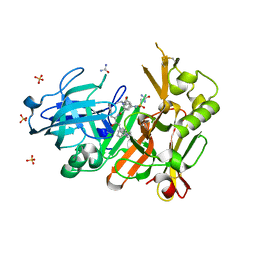 | | Crystal structure of bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[2-(1-benzylpiperidin-4-yl)ethyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide, SULFATE ION, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-05-25 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
To be Published, 2012
|
|
4F8Y
 
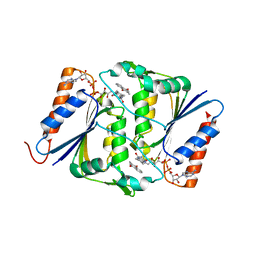 | |
3C6R
 
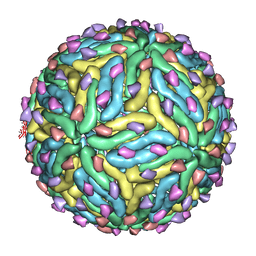 | | Low pH Immature Dengue Virus | | Descriptor: | Envelope protein, Peptide pr | | Authors: | Yu, I, Zhang, W, Holdway, H.A, Li, L, Kostyuchenko, V.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Structure of the immature dengue virus at low pH primes proteolytic maturation
Science, 319, 2008
|
|
3F9G
 
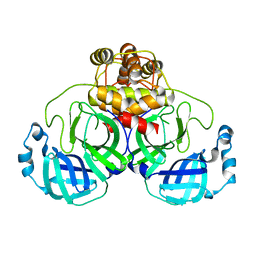 | |
3F9F
 
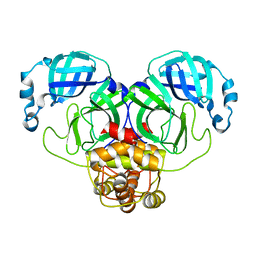 | |
3F9H
 
 | |
3F9E
 
 | |
2LH8
 
 | | Syrian hamster prion protein with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Major prion protein | | Authors: | Perez-Pineiro, R, Bjorndahl, T.C, Berjanskii, M, Hau, D, Li, L, Huang, A, Lee, R, Gibbs, E, Ladner, C, Wei Dong, Y, Abera, A, Cashman, N.R, Wishart, D. | | Deposit date: | 2011-08-05 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The prion protein binds thiamine.
Febs J., 278, 2011
|
|
2L6I
 
 | |
3G7F
 
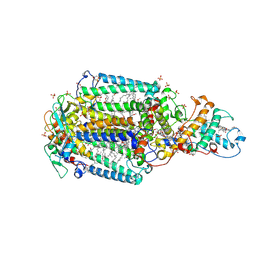 | | Crystal structure of Blastochloris viridis heterodimer mutant reaction center | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Ponomarenko, N.S, Li, L, Tereshko, V, Ismagilov, R.F, Norris Jr, J.R. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and spectropotentiometric analysis of Blastochloris viridis heterodimer mutant reaction center
Biochim.Biophys.Acta, 1788, 2009
|
|
3O4O
 
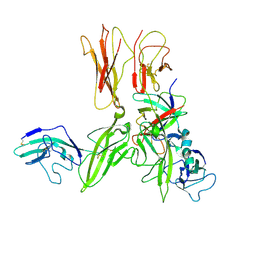 | | Crystal structure of an Interleukin-1 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, ... | | Authors: | Wang, X.Q, Wang, D.L, Zhang, S.Y, Li, L, Liu, X, Mei, K.R. | | Deposit date: | 2010-07-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the assembly and activation of IL-1beta with its receptors
Nat.Immunol., 11, 2010
|
|
3RT3
 
 | |
3II9
 
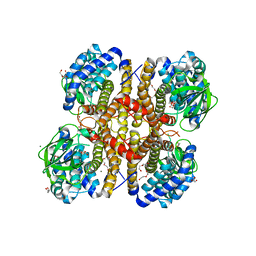 | | Crystal structure of glutaryl-coa dehydrogenase from Burkholderia pseudomallei at 1.73 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutaryl-CoA dehydrogenase, ... | | Authors: | Ismagilov, R.F, Li, L, Du, W.B, Staker, B, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-07-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | User-loaded SlipChip for equipment-free multiplexed nanoliter-scale experiments.
J.Am.Chem.Soc., 132, 2010
|
|
3L8R
 
 | | The crystal structure of PtcA from S. mutans | | Descriptor: | Putative PTS system, cellobiose-specific IIA component | | Authors: | Lei, J, Liu, X, Li, L. | | Deposit date: | 2010-01-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PtcA from Streptococcus mutans
To be Published
|
|
4MSR
 
 | | RNA 10mer duplex with six 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with six 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MS9
 
 | | Native RNA-10mer Structure: ccggcgccgg | | Descriptor: | Native RNA duplex 10mer, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MSB
 
 | | RNA 10mer duplex with two 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with two 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
