5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
2RI0
 
 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucosamine-6-phosphate deaminase, SODIUM ION | | Authors: | Li, D, Liu, C, Li, L.F, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3EXT
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | MAGNESIUM ION, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Liu, X, Li, G.L, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
7BRC
 
 | | Crystal structure of the TMK3 LRR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-like kinase TMK3 | | Authors: | Chen, H, Kong, Y.Q, Chen, J, Li, L, Li, X.S, Yu, F, Ming, Z.H. | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the extracellular domain of the receptor-like kinase TMK3 from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4E2A
 
 | |
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
6SCX
 
 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | Authors: | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | Deposit date: | 2019-07-25 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
6LKC
 
 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
3NSQ
 
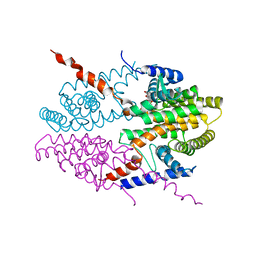 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | Descriptor: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3NSP
 
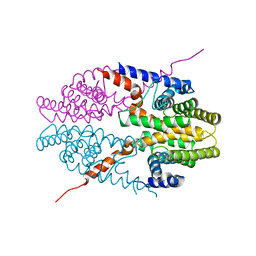 | | Crystal structure of tetrameric RXRalpha-LBD | | Descriptor: | Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
6J3P
 
 | | Crystal structure of the human GCN5 bromodomain in complex with compound (R,R)-36n | | Descriptor: | 2-{[(3R,5R)-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-methylpiperidin-3-yl]amino}-3-methyl-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2A | | Authors: | Huang, L.Y, Li, H, Niu, L, Wu, C.Y, Yu, Y.M, Li, L.L, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
8FZD
 
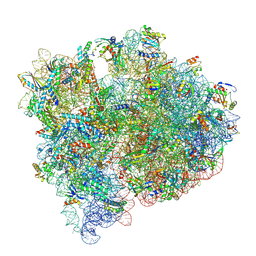 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZE
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZF
 
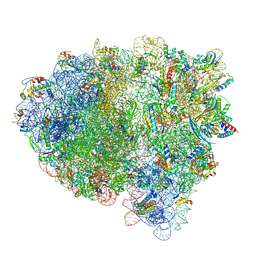 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZI
 
 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZH
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZJ
 
 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZG
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | Deposit date: | 2023-01-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
3H6X
 
 | |
6XKD
 
 | | Structure of ligand-bound mouse cGAMP hydrolase ENPP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Li, L. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Aided Development of Small-Molecule Inhibitors of ENPP1, the Extracellular Phosphodiesterase of the Immunotransmitter cGAMP.
Cell Chem Biol, 27, 2020
|
|
