4DV9
 
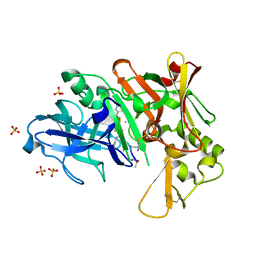 | | Crystal structure of BACE1 with its inhibitor | | 分子名称: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | 著者 | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | 登録日 | 2012-02-23 | | 公開日 | 2013-01-16 | | 最終更新日 | 2021-09-15 | | 実験手法 | X-RAY DIFFRACTION (2.076 Å) | | 主引用文献 | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
8KFO
 
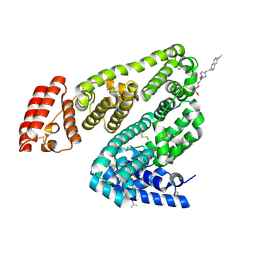 | | Crystal structure of BSA in complex with B3 | | 分子名称: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | 著者 | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | 登録日 | 2023-08-16 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | 分子名称: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | 著者 | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | 登録日 | 2008-12-09 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
3FHQ
 
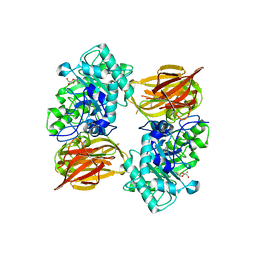 | | Structure of endo-beta-N-acetylglucosaminidase A | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | 著者 | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | 登録日 | 2008-12-10 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.452 Å) | | 主引用文献 | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
3NSQ
 
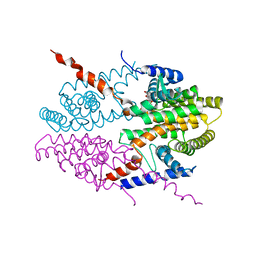 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | 分子名称: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | 著者 | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3NSP
 
 | | Crystal structure of tetrameric RXRalpha-LBD | | 分子名称: | Retinoid X receptor, alpha | | 著者 | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
7M2P
 
 | |
3H6X
 
 | |
8FZF
 
 | | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZE
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZI
 
 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZJ
 
 | | Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZH
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZD
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
8FZG
 
 | | Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rybak, M.Y, Li, L, Lin, J, Gagnon, M.G. | | 登録日 | 2023-01-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The ribosome termination complex remodels release factor RF3 and ejects GDP.
Nat.Struct.Mol.Biol., 2024
|
|
6SCX
 
 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | 著者 | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | 登録日 | 2019-07-25 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
4E2A
 
 | |
3DEZ
 
 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | 分子名称: | Orotate phosphoribosyltransferase, SULFATE ION | | 著者 | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | 登録日 | 2008-06-11 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2G0I
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | 分子名称: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, hypothetical protein SMU.848 | | 著者 | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | 登録日 | 2006-02-13 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
2HCU
 
 | | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans | | 分子名称: | 3-isopropylmalate dehydratase small subunit, SULFATE ION | | 著者 | Gao, Z.Q, Hou, H.F, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | 登録日 | 2006-06-19 | | 公開日 | 2006-07-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure Of Smu.1381 (or LeuD) from Streptococcus Mutans
To be Published
|
|
7MW8
 
 | |
2G0J
 
 | | Crystal structure of SMU.848 from Streptococcus mutans | | 分子名称: | hypothetical protein SMU.848 | | 著者 | Hou, H.-F, Gao, Z.-Q, Li, L.-F, Liang, Y.-H, Su, X.-D, Dong, Y.-H. | | 登録日 | 2006-02-13 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of SMU.848 from Streptococcus mutans
To be Published
|
|
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | 分子名称: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | 著者 | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | 登録日 | 2019-01-05 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
6CFF
 
 | | Stimulator of Interferon Genes Human | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon genes protein | | 著者 | Fernandez, D, Li, L, Ergun, S.L. | | 登録日 | 2018-02-14 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition.
Cell, 178, 2019
|
|
3UQP
 
 | | Crystal structure of Bace1 with its inhibitor | | 分子名称: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | 著者 | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | 登録日 | 2011-11-21 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
