7TYK
 
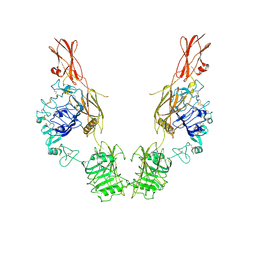 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | 分子名称: | Insulin receptor-related protein | | 著者 | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
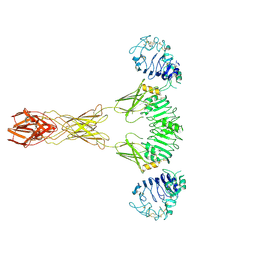 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | 分子名称: | Insulin receptor-related protein | | 著者 | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | 分子名称: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J, Zhu, R, Pei, Y. | | 登録日 | 2021-03-23 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-04-06 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-03-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7ENJ
 
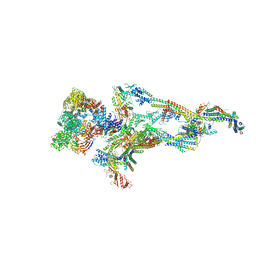 | | Human Mediator (deletion of MED1-IDR) in a Tail-bent conformation (MED-B) | | 分子名称: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, ... | | 著者 | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | 登録日 | 2021-04-17 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | 登録日 | 2021-04-16 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (4.07 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
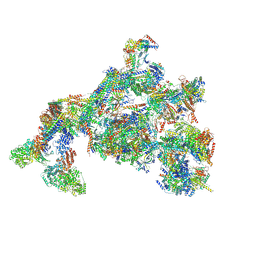 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | 分子名称: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | 登録日 | 2021-04-16 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (4.13 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7EMF
 
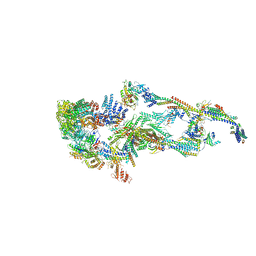 | | Human Mediator (deletion of MED1-IDR) in a Tail-extended conformation | | 分子名称: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 16, Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, ... | | 著者 | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | 登録日 | 2021-04-13 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7F4G
 
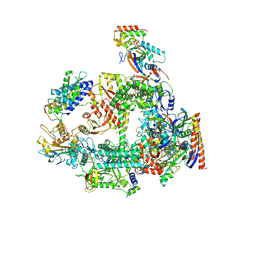 | | Structure of RPAP2-bound RNA polymerase II | | 分子名称: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | 著者 | Chen, X, Qi, Y, Wang, X, Li, J, Zhao, D, Xu, Y. | | 登録日 | 2021-06-18 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | RPAP2 regulates a transcription initiation checkpoint by inhibiting assembly of pre-initiation complex.
Cell Rep, 39, 2022
|
|
7CBY
 
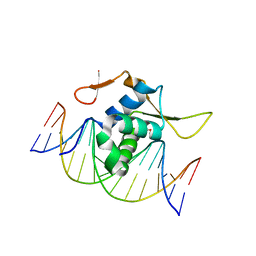 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | 分子名称: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | 著者 | Dai, S.Y, Li, J, Chen, Y.H. | | 登録日 | 2020-06-15 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.646 Å) | | 主引用文献 | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
7FJ2
 
 | |
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Liu, L, Li, J. | | 登録日 | 2021-06-05 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7X8L
 
 | |
4DZY
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with ADP | | 分子名称: | (S)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-03-01 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7CIP
 
 | | Microbial Hormone-sensitive lipase E53 wild type | | 分子名称: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | 著者 | Yang, X, Li, Z, Xu, X, Li, J. | | 登録日 | 2020-07-08 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Microbial Hormone-sensitive lipase E53 wild type
To Be Published
|
|
4H7Q
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase in complex with alpha-ketoisocaproic acid and ADP | | 分子名称: | 2-OXO-4-METHYLPENTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H85
 
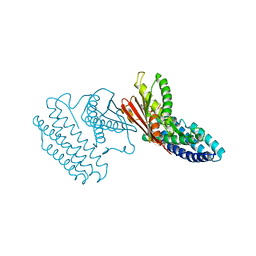 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-alpha-chloroisocaproate complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALPHA-CHLOROISOCAPROIC ACID, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TZ5
 
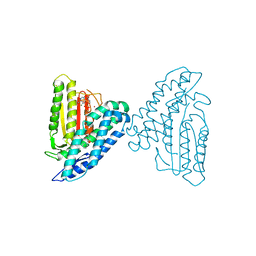 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex with ADP | | 分子名称: | 4-PHENYL-BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-27 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TZ2
 
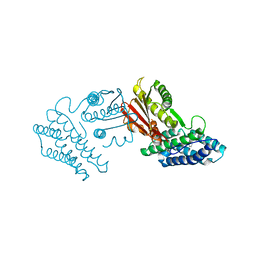 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/phenylbutyrate complex | | 分子名称: | 4-PHENYL-BUTANOIC ACID, [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-26 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4H81
 
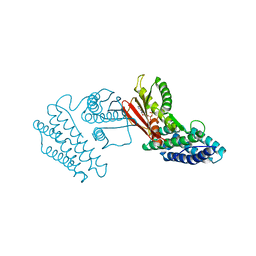 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(R)-2-chloro-3-phenylpropanoic acid complex with ADP | | 分子名称: | (2R)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2012-09-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TZ4
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex with ADP | | 分子名称: | (2S)-2-chloro-4-methylpentanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-27 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TZ0
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex | | 分子名称: | (2S)-2-chloro-4-methylpentanoic acid, [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-26 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7XOF
 
 | |
