3FP2
 
 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
3FP3
 
 | | Crystal structure of Tom71 | | Descriptor: | CHLORIDE ION, SULFATE ION, TPR repeat-containing protein YHR117W | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
6PYH
 
 | | Cryo-EM structure of full-length IGF1R-IGF1 complex. Only the extracellular region of the complex is resolved. | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Choi, E, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the activation of type 1 insulin-like growth factor receptor.
Nat Commun, 10, 2019
|
|
7CFU
 
 | |
3FP4
 
 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY4
 
 | | GajA tetramer with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | | tetramer Gabija with ATP (local refinement) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, MAGNESIUM ION | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Tetramer Gabija with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X51
 
 | | Cryo-EM structure of Gabija GajA in complex with DNA(focused refinement) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6WLG
 
 | | Ints3 C-terminal Domain | | Descriptor: | Integrator complex subunit 3 | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Structural basis for multifunctional roles of human Ints3 C-terminal domain.
J.Biol.Chem., 296, 2020
|
|
6WQX
 
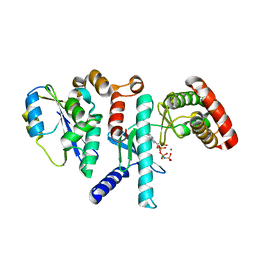 | | Human PRPK-TPRKB complex | | Descriptor: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
7CDB
 
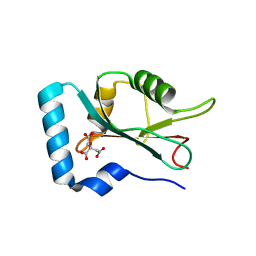 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | Descriptor: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | Authors: | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
8UOZ
 
 | | EmrE structure in the TPP-bound state (WT/E14Q heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE, TETRAPHENYLPHOSPHONIUM | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
5DWD
 
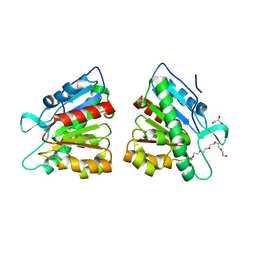 | | Crystal structure of esterase PE8 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Esterase, GLYCEROL | | Authors: | Li, J, Huang, J. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of esterase PE8
To Be Published
|
|
5HC4
 
 | | Structure of esterase Est22 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
4LWE
 
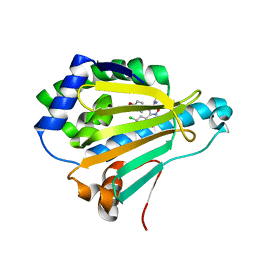 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWI
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | Authors: | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C2Z
 
 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | Descriptor: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Li, J, Zhu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
5HC0
 
 | | Structure of esterase Est22 with p-nitrophenol | | Descriptor: | ACETIC ACID, GLYCEROL, Lipolytic enzyme, ... | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC5
 
 | |
5HC3
 
 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC2
 
 | |
4LWF
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ3 | | Descriptor: | 4-(5-amino-1,2-oxazol-3-yl)-6-(propan-2-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
