7X8L
 
 | |
8DSV
 
 | | The structure of NicA2 in complex with N-methylmyosmine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent oxidoreductase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
8DQ7
 
 | |
8DQ8
 
 | | The structure of NicA2 variant F104L/A107T/S146I/G317D/H368R/L449V/N462S in complex with N-methylmyosmine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 1,2-ETHANEDIOL, Amine oxidase, ... | | Authors: | Wu, K, Dulchavsky, M, Li, J, Bardwell, J.C.A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed evolution unlocks oxygen reactivity for a nicotine-degrading flavoenzyme.
Nat.Chem.Biol., 19, 2023
|
|
5GVA
 
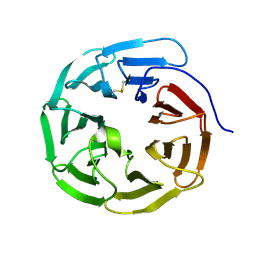 | | WD40 domain of human AND-1 | | Descriptor: | WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Guan, C.C, Li, J. | | Deposit date: | 2016-09-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome
J. Biol. Chem., 292, 2017
|
|
6J72
 
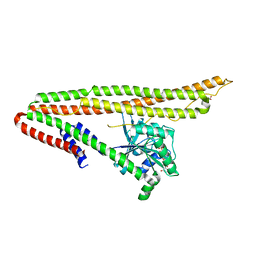 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
5GVB
 
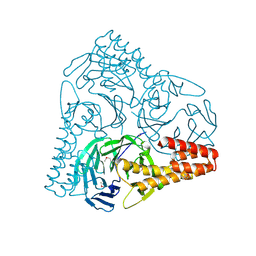 | | SepB domain of human AND-1 | | Descriptor: | WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Guan, C.C, Li, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome.
J. Biol. Chem., 292, 2017
|
|
4PT1
 
 | |
1TR4
 
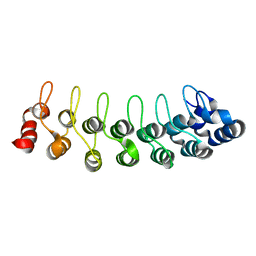 | | Solution structure of human oncogenic protein gankyrin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10 | | Authors: | Yuan, C, Li, J, Mahajan, A, Poi, M.J, Byeon, I.J, Tsai, M.D. | | Deposit date: | 2004-06-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship.
Biochemistry, 43, 2004
|
|
4NXB
 
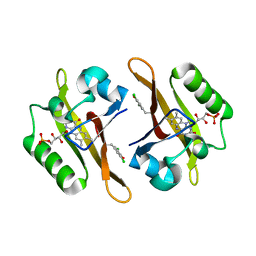 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
6N43
 
 | |
8H91
 
 | |
6N42
 
 | |
8HQH
 
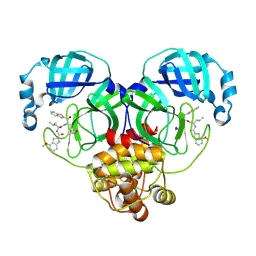 | |
8HQJ
 
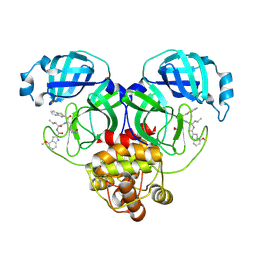 | |
8HQF
 
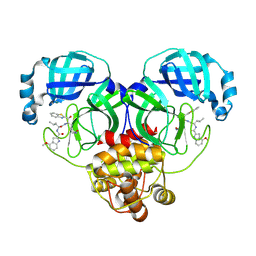 | |
4NXE
 
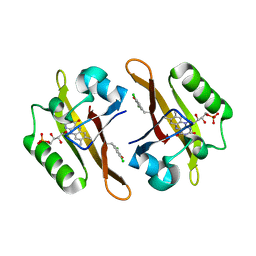 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
1BU9
 
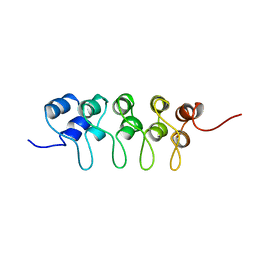 | | SOLUTION STRUCTURE OF P18-INK4C, 21 STRUCTURES | | Descriptor: | PROTEIN (CYCLIN-DEPENDENT KINASE 6 INHIBITOR) | | Authors: | Byeon, I.-J.L, Li, J, Tsai, M.-D. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A.
Biochemistry, 38, 1999
|
|
8FIA
 
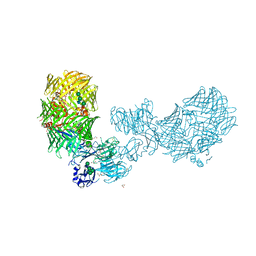 | | The structure of fly Teneurin self assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bandekar, S.J, Li, J, Arac, D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of fly Teneurin-m reveals an asymmetric self-assembly that allows expansion into zippers.
Embo Rep., 24, 2023
|
|
4JVG
 
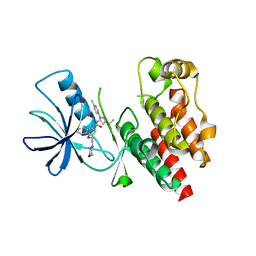 | | B-Raf Kinase in Complex with Birb796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Thevakumaran, N, Gavory, G, Li, J, Padeganeh, A, Guiral, S, Duchaine, J, Mao, D.Y.L, Bouvier, M, Sicheri, F, Therrien, M. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Inhibitors that stabilize a closed RAF kinase domain conformation induce dimerization.
Nat.Chem.Biol., 9, 2013
|
|
8HVL
 
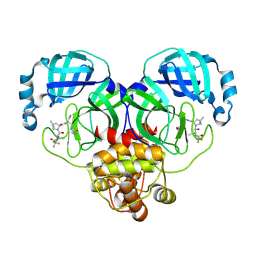 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF07321332
To Be Published
|
|
8HVO
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with PF07321332
To Be Published
|
|
8HVK
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
G15S mutant in complex with PF07321332
To Be Published
|
|
8HZR
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
1MI1
 
 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
