2AVF
 
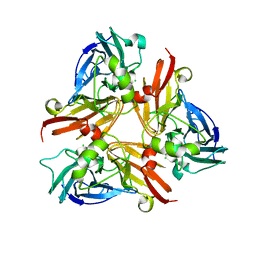 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
2F6N
 
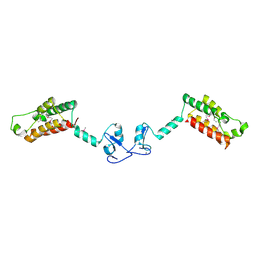 | |
2YS1
 
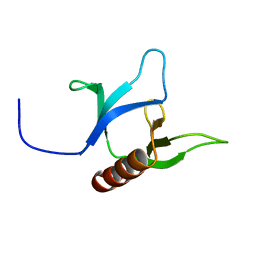 | | Solution structure of the PH domain of Dynamin-2 from human | | Descriptor: | Dynamin-2 | | Authors: | Li, H, Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Dynamin-2 from human
To be Published
|
|
2YRY
 
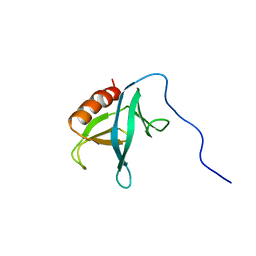 | | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human | | Descriptor: | Pleckstrin homology domain-containing family A member 6 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Pleckstrin homology domain-containing family A member 6 from human
To be Published
|
|
2YS3
 
 | | Solution structure of the PH domain of Kindlin-3 from human | | Descriptor: | Unc-112-related protein 2 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Kindlin-3 from human
To be Published
|
|
2YSZ
 
 | | Solution structure of the chimera of the C-terminal PID domain of Fe65L and the C-terminal tail peptide of APP | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2 and Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
4OIM
 
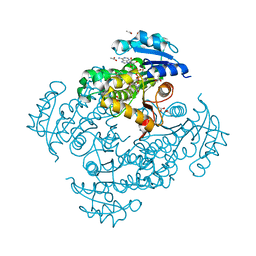 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
4OHU
 
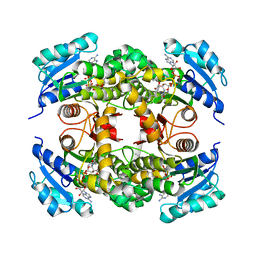 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
5CP4
 
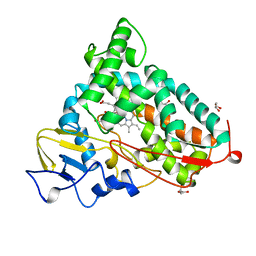 | | CRYOGENIC STRUCTURE OF P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-05-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect.
Biochemistry, 37, 1998
|
|
3E9D
 
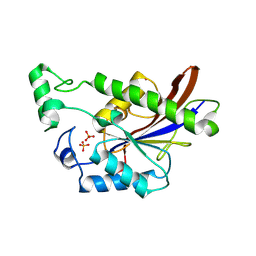 | | Structure of full-length TIGAR from Danio rerio | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Zgc:56074 | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2008-08-21 | | Release date: | 2008-12-16 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TIGAR (TP53 induced glycolysis and apoptosis regulator) is a fructose-2,6- and fructose-1,6-bisphosphatase
To be Published
|
|
3D1D
 
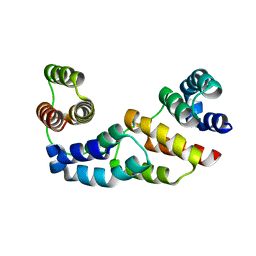 | | Hexagonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
8P30
 
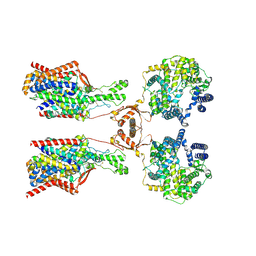 | | Structure of human SIT1:ACE2 complex (open PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Y
 
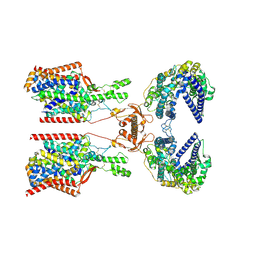 | | Structure of human SIT1:ACE2 complex (closed PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P31
 
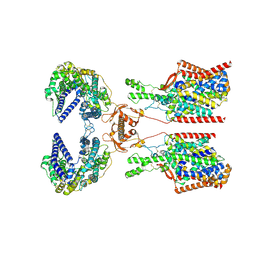 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
5HK8
 
 | |
3FD2
 
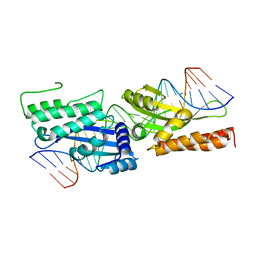 | | Crystal structure of mMsoI/DNA complex with calcium | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Li, H, Monnat, R.J. | | Deposit date: | 2008-11-24 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins.
Nucleic Acids Res., 37, 2009
|
|
3NMU
 
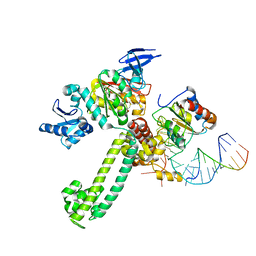 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-06-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
2YT0
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Tochio, N, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2YS5
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2YT2
 
 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | Fibroblast growth factor receptor substrate 3 and ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
3NVI
 
 | | Structure of N-terminal truncated Nop56/58 bound with L7Ae and box C/D RNA | | Descriptor: | 50S ribosomal protein L7Ae, NOP5/NOP56 related protein, RNA (5'-R(*CP*UP*CP*UP*GP*AP*CP*CP*GP*AP*AP*AP*GP*GP*CP*GP*UP*GP*AP*UP*GP*AP*GP*C)-3') | | Authors: | Li, H, Xue, S, Wang, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
2YS4
 
 | | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human | | Descriptor: | Hydrocephalus-inducing protein homolog | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal PapD-like domain of HYDIN protein from human
To be Published
|
|
2YT1
 
 | | Solution structure of the chimera of the C-terminal tail peptide of APP and the C-terminal PID domain of Fe65L | | Descriptor: | Amyloid beta A4 protein and Amyloid beta A4 precursor protein-binding family B member 2 | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
1WGU
 
 | | Solution Structure of the C-terminal Phosphotyrosine Interaction Domain of APBB2 from Mouse | | Descriptor: | amyloid beta (A4) precursor protein-binding, family B, member 2 | | Authors: | Li, H, Hayashi, F, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
