8J21
 
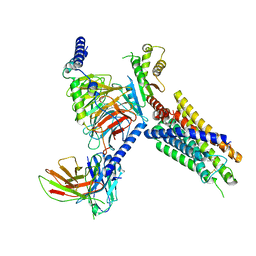 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J23
 
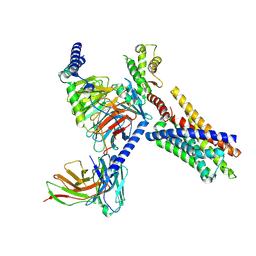 | | Cryo-EM structure of FFAR2 complex in apo state | | Descriptor: | Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short chain fatty acid receptors FFAR2 and FFAR3
To Be Published
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | Descriptor: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4XKL
 
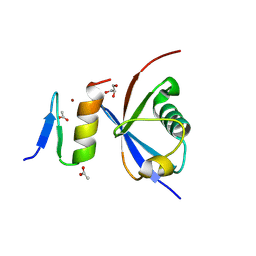 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
7R1U
 
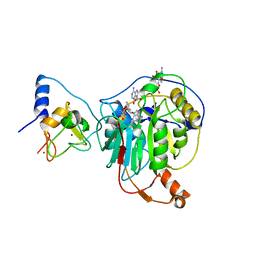 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
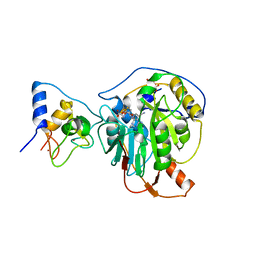 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
9ATP
 
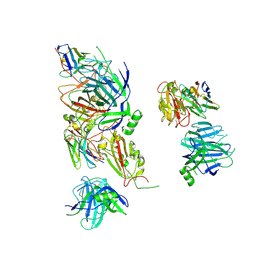 | |
9ATO
 
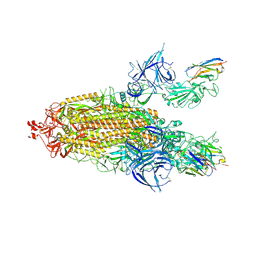 | | XBB.1.5 spike/Nanosota-3C complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3C, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2024-02-27 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of XBB.1.5 spike/Nanosota-3C complex
To Be Published
|
|
8QN0
 
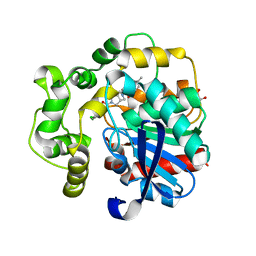 | | Soluble epoxide hydrolase in complex with RK3 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-5-(4-methylphenyl)sulfonyl-1,3,3~{a},4,6,6~{a}-hexahydropyrrolo[3,4-c]pyrrole-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
8QMZ
 
 | | Soluble epoxide hydrolase in complex with RK4 | | Descriptor: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-25 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
3T7J
 
 | | Crystal structure of Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7I
 
 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7K
 
 | | Complex structure of Rtt107p and phosphorylated histone H2A | | Descriptor: | Histone H2A.1, Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
8VQR
 
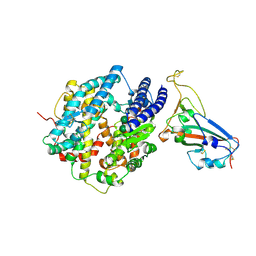 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
6VSJ
 
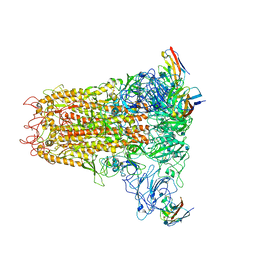 | | Cryo-electron microscopy structure of mouse coronavirus spike protein complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, Spike glycoprotein | | Authors: | Shang, J, Wan, Y.S, Liu, C, Yount, B, Gully, K, Yang, Y, Auerbach, A, Peng, G.Q, Baric, R, Li, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry.
Plos Pathog., 16, 2020
|
|
6VW1
 
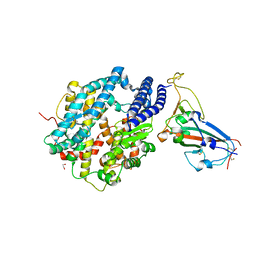 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | Deposit date: | 2020-02-18 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
7TGX
 
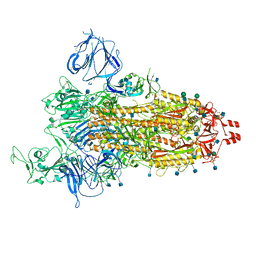 | | Prototypic SARS-CoV-2 G614 spike (open form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGY
 
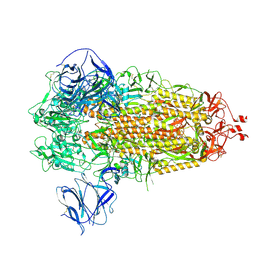 | | Prototypic SARS-CoV-2 G614 spike (closed form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7TGW
 
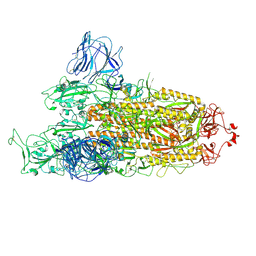 | | Omicron spike at 3.0 A (open form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ye, G, Liu, B, Li, F. | | Deposit date: | 2022-01-09 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain.
Nat Commun, 13, 2022
|
|
7U0N
 
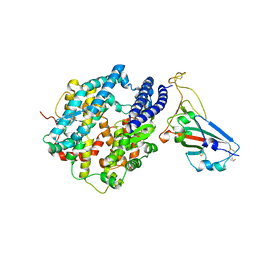 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
4R7A
 
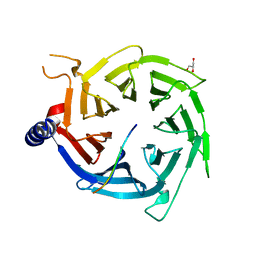 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
4X0V
 
 | | Structure of a GH5 family lichenase from Caldicellulosiruptor sp. F32 | | Descriptor: | Beta-1,3-1,4-glucanase | | Authors: | Meng, D, Liu, X, Wang, X, Li, F, Feng, Y. | | Deposit date: | 2014-11-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structural Insights into the Substrate Specificity of a Glycoside Hydrolase Family 5 Lichenase from Caldicellulosiruptor sp. F32
Biochem. J., 2017
|
|
3V43
 
 | | Crystal structure of MOZ | | Descriptor: | ACETATE ION, Histone H3.1, Histone acetyltransferase KAT6A, ... | | Authors: | Qiu, Y, Li, F. | | Deposit date: | 2011-12-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription
Genes Dev., 26, 2012
|
|
5J2Y
 
 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
8THF
 
 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
