7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4QFL
 
 | |
5EQN
 
 | | Structure of phosphonate hydroxylase | | Descriptor: | FrbJ, MAGNESIUM ION | | Authors: | Li, C, Hu, Y, Zhang, H. | | Deposit date: | 2015-11-13 | | Release date: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a phosphonate hydroxylase with an access tunnel at the back of the active site.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7CQW
 
 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQQ
 
 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4QFN
 
 | |
4QFP
 
 | |
7CQL
 
 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4KRW
 
 | |
7WDW
 
 | | DsyB in complex with SAH and MTHB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-methylsulfanyl-2-oxidanyl-butanoic acid, DSYB, ... | | Authors: | Li, C.Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mechanistic insights into the key marine dimethylsulfoniopropionate synthesis enzyme DsyB/DSYB.
Mlife, 2022
|
|
7WDQ
 
 | | DsyB in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, SAM-dependent MTHB methyltransferase | | Authors: | Li, C.Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic insights into the key marine dimethylsulfoniopropionate synthesis enzyme DsyB/DSYB.
Mlife, 2022
|
|
7YLE
 
 | | RnDmpX in complex with DMSP | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, Glycine betaine/proline ABC transporter, periplasmic glycinebetaine/proline-binding protein | | Authors: | Li, C.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria.
Isme J, 17, 2023
|
|
7D4N
 
 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 20 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4M
 
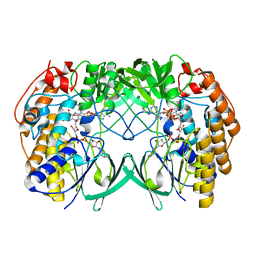 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 5 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4K
 
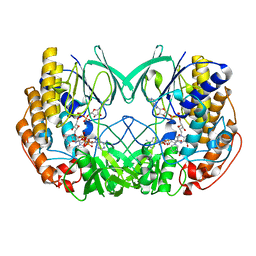 | | Crystal structure of Tmm from Pelagibacter sp. strain HTCC7211 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
8OS5
 
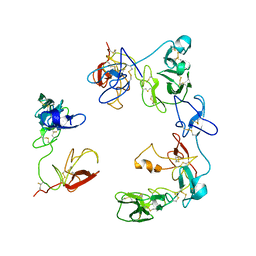 | | Crystal structure of the Factor XII heavy chain reveals an interlocking dimer with a FnII to kringle domain interaction | | Descriptor: | Coagulation factor XII-Mie | | Authors: | Li, C, Saleem, M, Kaira, B.G, Brown, A, Wilson, C, Philippou, H, Emsley, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Factor XII and kininogen asymmetric assembly with gC1qR/C1QBP/P32 is governed by allostery.
Blood, 136, 2020
|
|
6K9X
 
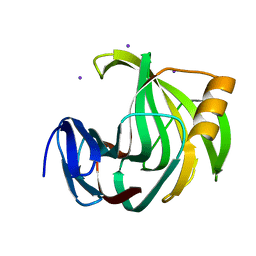 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism
Acta Crystallographica Section D-Biological Crystallography, D70, 2014
|
|
6JZP
 
 | |
4F5A
 
 | | Triple mutant Src SH2 domain bound to phosphate ion | | Descriptor: | PHOSPHATE ION, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F5B
 
 | | Triple mutant Src SH2 domain bound to phosphotyrosine | | Descriptor: | O-PHOSPHOTYROSINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4F59
 
 | | Triple mutant Src SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kaneko, T, Huang, H, Cao, X, Li, C, Voss, C, Sidhu, S.S, Li, S.S. | | Deposit date: | 2012-05-12 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Superbinder SH2 Domains Act as Antagonists of Cell Signaling.
Sci.Signal., 5, 2012
|
|
4MZ4
 
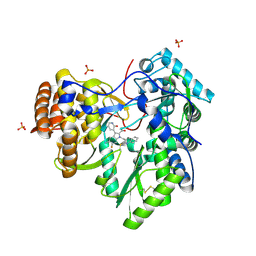 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4XZ6
 
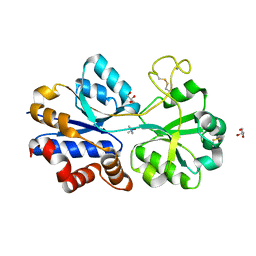 | | TmoX in complex with TMAO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
