1ZNS
 
 | | Crystal structure of N-ColE7/12-bp DNA/Zn complex | | Descriptor: | 5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*CP*G)-3', Colicin E7, ZINC ION | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
2VMJ
 
 | | Type 1 Copper-Binding Loop of Nitrite Reductase mutant: 130- CAPEGMVPWHVVSGM-144 to 130-CTPHPFM-136 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Sato, K, Firbank, S.J, Li, C, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Importance of the Long Type 1 Copper-Binding Loop of Nitrite Reductase for Structure and Function.
Chemistry, 14, 2008
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
1KBK
 
 | | Mechanistic Analyses of Catalysis in Human Pancreatic Alpha-Amylase: Detailed Kinetic and Structural Studies of Mutants of Three Conserved Carboxylic Acids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Rydberg, E.H, Li, C, Maurus, R, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids.
Biochemistry, 41, 2002
|
|
1KBB
 
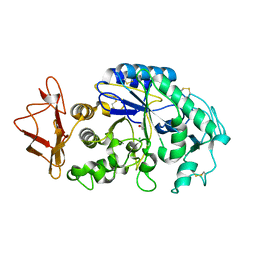 | | Mechanistic Analyses of Catalysis in Human Pancreatic alpha-Amylase: Detailed Kinetic and Structural Studies of Mutants of Three Conserved Carboxylic Acids | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Rydberg, E.H, Li, C, Maurus, R, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-05 | | Release date: | 2002-04-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic analyses of catalysis in human pancreatic alpha-amylase: detailed kinetic and structural studies of mutants of three conserved carboxylic acids.
Biochemistry, 41, 2002
|
|
4J4V
 
 | | Pentameric SFTSVN with suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
8IF5
 
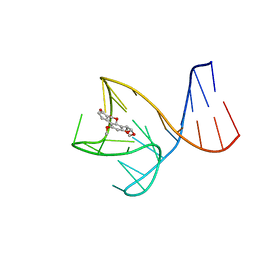 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
4J4S
 
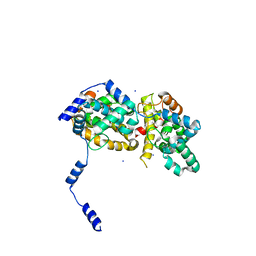 | | Triple mutant SFTAVN | | Descriptor: | Nucleocapsid protein, SODIUM ION | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
4J4X
 
 | | Crystal structure of GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4Y
 
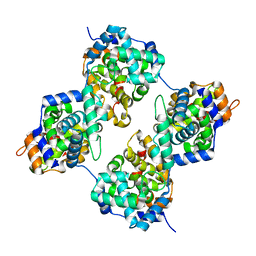 | | Triple mutant GraVN | | Descriptor: | NP protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4U
 
 | | Pentamer SFTSVN | | Descriptor: | Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
1ZNV
 
 | | How a His-metal finger endonuclease ColE7 binds and cleaves DNA with a transition metal ion cofactor | | Descriptor: | Colicin E7, Colicin E7 immunity protein, NICKEL (II) ION, ... | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
8I4T
 
 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
1QX3
 
 | | Conformational restrictions in the active site of unliganded human caspase-3 | | Descriptor: | Apopain | | Authors: | Ni, C.-Z, Li, C, Wu, J.C, Spada, A.P, Ely, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational restrictions in the active site of unliganded human caspase-3
J.MOL.RECOG., 16, 2003
|
|
5GSN
 
 | | Tmm in complex with methimazole | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Structural mechanism for bacterial oxidation of oceanic trimethylamine into trimethylamine N-oxide
Mol. Microbiol., 103, 2017
|
|
5TYT
 
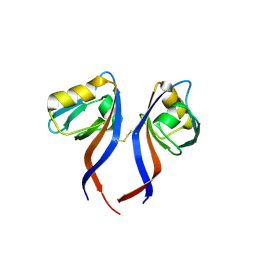 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
1YSG
 
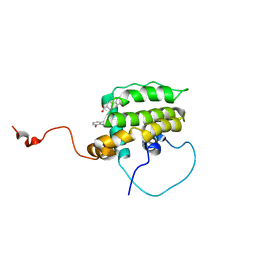 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1ZMS
 
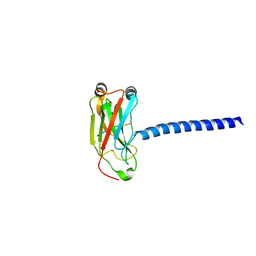 | | LMP1 Protein binds to TRAF3 as a structural CD40 | | Descriptor: | Latent membrane protein 1, TNF receptor associated factor 3 | | Authors: | Wu, S.D, Xie, P, Welsh, K, Li, C, Ni, C.-Z, Zhu, X, Reed, J.C, Satterthwait, A.C, Bishop, G.A, Ely, K.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | LMP1 protein from the Epstein Barr virus is a structural CD40 decoy in B lymphocytes fro binding to TRAF3
J.Biol.Chem., 39, 2005
|
|
1ZYW
 
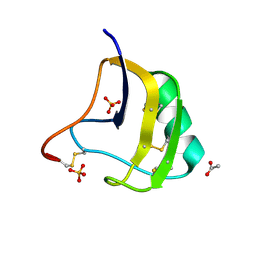 | | Crystal Structure Of Mutant K8DP9SR58KP60G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1JXS
 
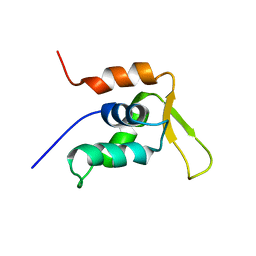 | | Solution Structure of the DNA-Binding Domain of Interleukin Enhancer Binding Factor | | Descriptor: | interleukin enhancer binding factor | | Authors: | Chuang, W.J, Liu, P.P, Li, C, Hsieh, Y.H, Chen, S.W, Chen, S.H, Jeng, W.Y. | | Deposit date: | 2001-09-08 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a)
PROTEINS: STRUCT.,FUNCT.,GENET., 49, 2002
|
|
4J4R
 
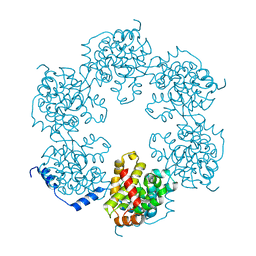 | | Hexameric SFTSVN | | Descriptor: | Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
4J4W
 
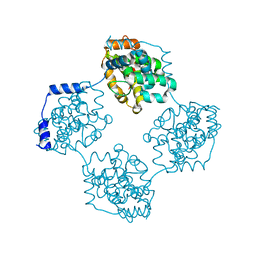 | | Crystal structure of BueVN | | Descriptor: | Nucleocapsid | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
