1BC1
 
 | | RECOMBINANT RAT ANNEXIN V, QUADRUPLE MUTANT (T72K, S144K, S228K, S303K) | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCW
 
 | | RECOMBINANT RAT ANNEXIN V, T72A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BC0
 
 | | RECOMBINANT RAT ANNEXIN V, W185A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCZ
 
 | | RECOMBINANT RAT ANNEXIN V, T72S MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCY
 
 | | RECOMBINANT RAT ANNEXIN V, T72K MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BC3
 
 | | RECOMBINANT RAT ANNEXIN V, TRIPLE MUTANT (T72K, S144K, S228K) | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
4M8V
 
 | |
2B0G
 
 | |
2AYM
 
 | |
4P5U
 
 | | Crystal structure of TatD | | Descriptor: | Tat-linked quality control protein TatD | | Authors: | Chen, Y, Li, C.-L, Hsiao, Y.-Y, Duh, Y, Yuan, H.S. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of TatD exonuclease in DNA repair.
Nucleic Acids Res., 42, 2014
|
|
5EOD
 
 | | Human Plasma Coagulation FXI with peptide LP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, LP2 | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
7CRD
 
 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
4M9O
 
 | |
5GGM
 
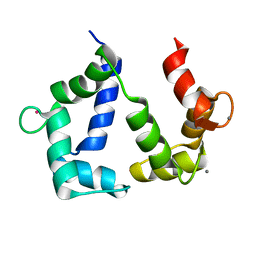 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
8OMV
 
 | |
7F77
 
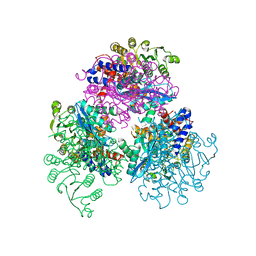 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | Descriptor: | Glutamate dehydrogenase | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F79
 
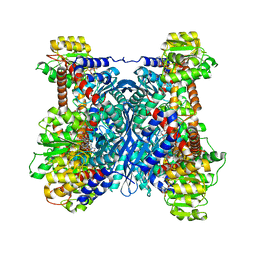 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | Authors: | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
5C70
 
 | | The structure of Aspergillus oryzae beta-glucuronidase | | Descriptor: | Glucuronidase | | Authors: | Sun, H.L, Lv, B, Huang, S, Sun, Q.F, Li, C, Jiang, T. | | Deposit date: | 2015-06-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enhancing the Thermostability of beta-Glucuronidase by Rationally Redesigning the Catalytic Domain Based on Sequence Alignment Strategy
Ind Eng Chem Res, 55, 2016
|
|
2I8T
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP-mannose complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GLYCEROL, ... | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
5EOK
 
 | | Human Plasma Coagulation Factor XI in complex with peptide P39 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XI, ... | | Authors: | Wong, S.S, Ostergaard, S, Hall, G, Li, C, Williams, P.M, Stennicke, H, Emsley, J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Blood, 127, 2016
|
|
2I8U
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
4Q8K
 
 | | Crystal structure of polysaccharide lyase family 18 aly-SJ02 P-CATD | | Descriptor: | Alginase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4Q8L
 
 | | Crystal structure of polysacchride lyase family 18 aly-SJ02 r-CATD | | Descriptor: | Alginase, CALCIUM ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4PQW
 
 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
