4Z44
 
 | | F454K Mutant of Tryptophan 7-halogenase PrnA | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Shepherd, S.A, Karthikeyan, C, Latham, J, Struck, A.-W, Thompson, M.L, Menon, B, Levy, C.W, Leys, D, Micklefield, J. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
3K8E
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
5A35
 
 | | Crystal structure of Glycine Cleavage Protein H-Like (GcvH-L) from Streptococcus pyogenes | | Descriptor: | GLYCINE CLEAVAGE SYSTEM H PROTEIN, PENTAETHYLENE GLYCOL | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
3EKF
 
 | |
3EKD
 
 | |
3F03
 
 | |
4JGI
 
 | | 1.5 Angstrom crystal structure of a novel cobalamin-binding protein from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CO-METHYLCOBALAMIN, Putative uncharacterized protein | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G48
 
 | | Structure of CYP121 in complex with 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3DG9
 
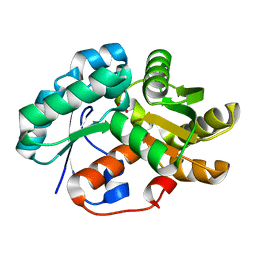 | | Crystal Structure of Malonate Decarboxylase from Bordatella bronchiseptica | | Descriptor: | Arylmalonate decarboxylase, PHOSPHATE ION | | Authors: | Okrasa, K, Levy, C, Baudendistel, N, Leys, D, Micklefield, J. | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of an Unusual Malonate Decarboxylase and Related Racemases.
Chemistry, 14, 2008
|
|
4J34
 
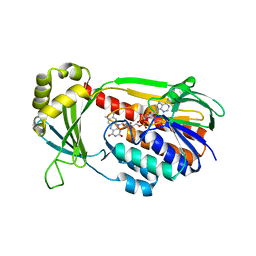 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4L2H
 
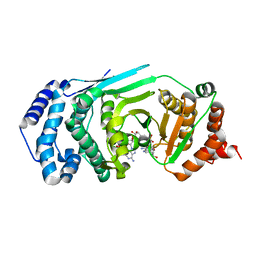 | | Structure of a catalytically inactive PARG in complex with a poly-ADP-ribose fragment | | Descriptor: | Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Brassington, A, Dunstan, M.S, Leys, D. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities.
Nat Commun, 4, 2013
|
|
2HJ4
 
 | |
3KRU
 
 | | Crystal Structure of the Thermostable Old Yellow Enzyme from Thermoanaerobacter pseudethanolicus E39 | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-11-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
3KRZ
 
 | | Crystal Structure of the Thermostable NADH4-bound old yellow enzyme from Thermoanaerobacter pseudethanolicus E39 | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-11-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
3LA5
 
 | |
3IA5
 
 | | Moritella profunda dihydrofolate reductase (DHFR) | | Descriptor: | Dihydrofolate reductase, PHOSPHATE ION | | Authors: | Hay, S, Evans, R.M, Levy, C, Wang, X, Loveridge, E.J, Leys, D, Allemann, R.K, Scrutton, N.S. | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Are the Catalytic Properties of Enzymes from Piezophilic Organisms Pressure Adapted?
Chembiochem, 10, 2009
|
|
3KX5
 
 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant F261E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
3KFT
 
 | | Crystal structure of Pentaerythritol Tetranitrate Reductase complex with 1,4,5,6-tetrahydro NADH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Pudney, C.R, Levy, C.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-10-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction.
J.Am.Chem.Soc., 131, 2009
|
|
3KX3
 
 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant L86E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
3KX4
 
 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant I401E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
2H6B
 
 | |
2HKR
 
 | |
2HJB
 
 | |
2HKM
 
 | |
2HXC
 
 | |
