6J8G
 
 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 9 subunit alpha, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
6J8H
 
 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 9 subunit alpha, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J6Q
 
 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6LGJ
 
 | | Crystal structure of an oxido-reductase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Lei, J, Yin, L. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an oxido-reductase
To be published
|
|
6LGK
 
 | |
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J8J
 
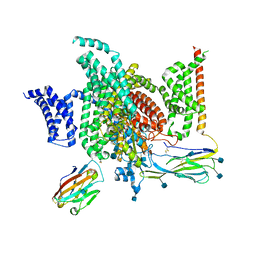 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 down) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
3D3F
 
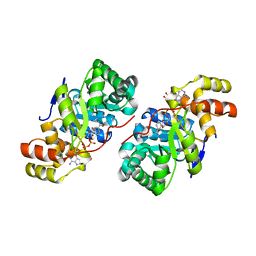 | |
3J3R
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine
J.Biol.Chem., 288, 2013
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
3J3T
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3F7J
 
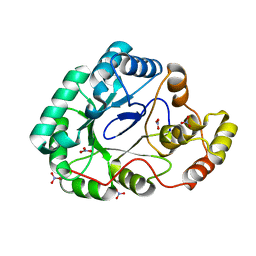 | | B.subtilis YvgN | | Descriptor: | NITRATE ION, POTASSIUM ION, YvgN protein | | Authors: | Zhou, Y.F, Lei, J, Liang, Y.H, Su, X.-D. | | Deposit date: | 2008-11-09 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of YvgN and YtbE from Bacillus subtilis
Protein Sci., 18, 2009
|
|
3J3U
 
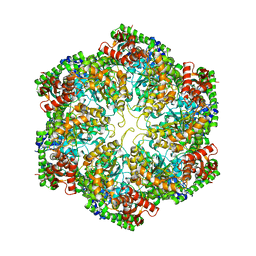 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J3S
 
 | | Structural dynamics of the MecA-ClpC complex revealed by cryo-EM | | Descriptor: | Adapter protein MecA 1, Negative regulator of genetic competence ClpC/MecB | | Authors: | Liu, J, Mei, Z, Li, N, Qi, Y, Xu, Y, Shi, Y, Wang, F, Lei, J, Gao, N. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural dynamics of the MecA-ClpC complex: a type II AAA+ protein unfolding machine.
J.Biol.Chem., 288, 2013
|
|
3J2G
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2E
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J28
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (12.9 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2D
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.700001 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J29
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2F
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (17.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2C
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA body domain, 16S rRNA head domain | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2H
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
