5LYV
 
 | | The crystal structure of 7SK 5'-hairpin - Osmium derivative | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
4FMO
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5LYU
 
 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
5LYS
 
 | | The crystal structure of 7SK 5'-hairpin - Gold derivative | | Descriptor: | 7SK RNA, GOLD ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
2X74
 
 | | Human foamy virus integrase - catalytic core. | | Descriptor: | INTEGRASE | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Legrand, P, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-11 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7ATH
 
 | | Crystal structure of UipA | | Descriptor: | UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
1OAO
 
 | | NiZn[Fe4S4] and NiNi[Fe4S4] clusters in closed and open alpha subunits of acetyl-CoA synthase/carbon monoxide dehydrogenase | | Descriptor: | ACETATE ION, BICARBONATE ION, CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Darnault, C, Volbeda, A, Kim, E.J, Legrand, P, Vernede, X, Lindahl, P.A, Fontecilla-Camps, J.C. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ni-Zn-[Fe4-S4] and Ni-Ni-[Fe4-S4] Clusters in Closed and Open Alpha Subunits of Acetyl-Coa Synthase/Carbon Monoxide Dehydrogenase
Nat.Struct.Biol., 10, 2003
|
|
7ATK
 
 | | Crystal structure of UipA in complex with Uranium | | Descriptor: | URANIUM ATOM, UipA, ZINC ION | | Authors: | Bremond, N, Gallois, N, Legrand, P, Chapon, V, Arnoux, P. | | Deposit date: | 2020-10-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Discovery and characterization of UipA, a uranium- and iron-binding PepSY protein involved in uranium tolerance by soil bacteria.
Isme J, 16, 2022
|
|
3UH8
 
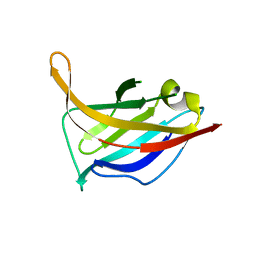 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OW3
 
 | | Crystal structure of a C-terminally truncated trimeric ectodomain of the Arabidopsis thaliana gamete fusion protein HAP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Fedry, J, Legrand, P, Rey, F.A, Krey, T. | | Deposit date: | 2017-08-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes.
PLoS Biol., 16, 2018
|
|
5EPV
 
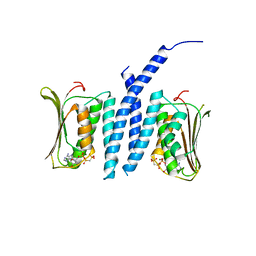 | | Histidine kinase domain from the LOV-HK blue-light receptor from Brucella abortus | | Descriptor: | Blue-light-activated histidine kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Rinaldi, J, Guimaraes, B.G, Legrand, P, Thompson, A, Paris, G, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the HWE Histidine Kinase Family: The Brucella Blue Light-Activated Histidine Kinase Domain.
J.Mol.Biol., 428, 2016
|
|
5OY9
 
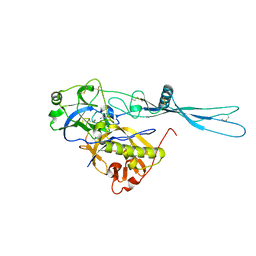 | | VSV G CR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albertini, A.A, Belot, L, Legrand, P, Gaudin, Y. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein.
Nat Commun, 9, 2018
|
|
2X78
 
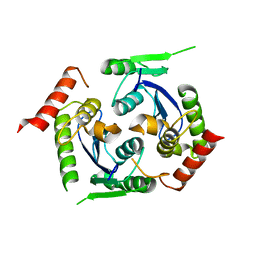 | | Human foamy virus integrase - catalytic core. | | Descriptor: | INTEGRASE | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Legrand, P, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6YRZ
 
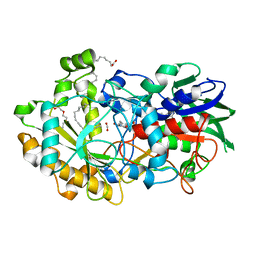 | | Crystal structure of FAP et pH 8.5 after illumination at 150K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, ... | | Authors: | Sorigue, D, Legrand, P, Blangy, S, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7QBU
 
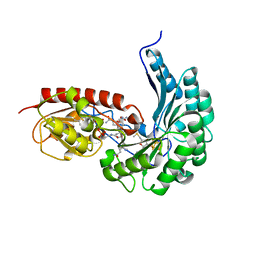 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBT
 
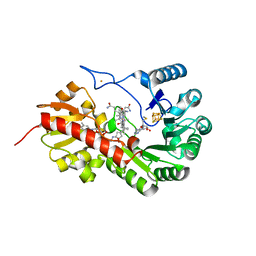 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, FE (III) ION, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBV
 
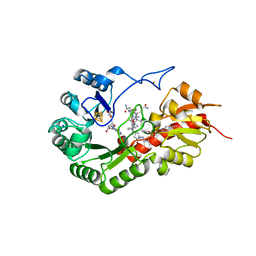 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7ANQ
 
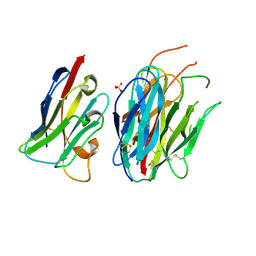 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
5LLU
 
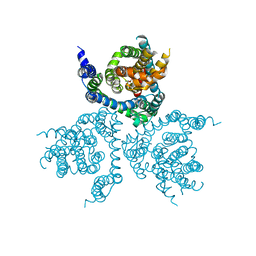 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
5LM4
 
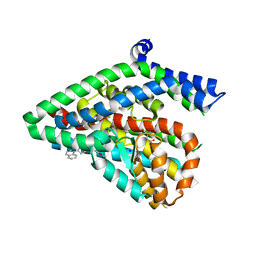 | | Structure of the thermostalilized EAAT1 cryst-II mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
5LLM
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-27 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
4HH6
 
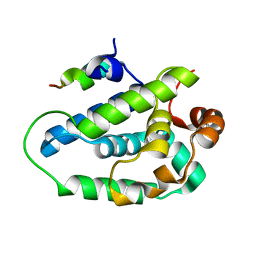 | | Peptide from EAEC T6SS Sci1 SciI protein | | Descriptor: | Peptide from EAEC T6SS Sci1 SciI protein, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
4HH5
 
 | | N-terminal domain (1-163) of ClpV1 ATPase from E.coli EAEC Sci1 T6SS. | | Descriptor: | BROMIDE ION, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
5M30
 
 | | Structure of TssK from T6SS EAEC in complex with nanobody nb18 | | Descriptor: | Anti-vesicular stomatitis virus N VHH, Type VI secretion protein | | Authors: | Nguyen, V.S, Cambillau, C, Spinelli, C, Desmyter, A, Legrand, P, Cascales, E. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-21 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
5M2Y
 
 | | Structure of TssK C-terminal domain from E. coli T6SS | | Descriptor: | TssK C | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A, Legrand, P. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
