5ZZ7
 
 | | Redox-sensing transcriptional repressor Rex | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Redox-sensing transcriptional repressor Rex 1 | | 著者 | Park, Y.W, Jang, Y.Y, Joo, H.K, Lee, J.Y. | | 登録日 | 2018-05-30 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Analysis of Redox-sensing Transcriptional Repressor Rex from Thermotoga maritima
Sci Rep, 8, 2018
|
|
3SMZ
 
 | |
4QIU
 
 | |
4J4J
 
 | |
4RS7
 
 | |
1BI8
 
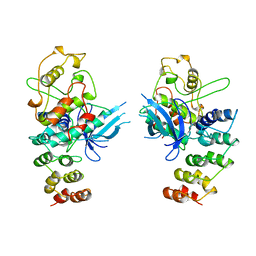 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | 分子名称: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | 著者 | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | 登録日 | 1998-06-22 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
3MMS
 
 | | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine | | 分子名称: | 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase, 9H-purine-6,8-diamine, GLYCEROL | | 著者 | Siu, K.K.W, Lee, J.E, Horvatin-Mrakovcic, C, Howell, P.L. | | 登録日 | 2010-04-20 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Streptococcus pneumoniae MTA/SAH nucleosidase in complex with 8-aminoadenine
TO BE PUBLISHED
|
|
3CQA
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ala and Lys101Ala | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-02 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRI
 
 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser, Glu82Asn and Lys101Ala | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-07 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3CRH
 
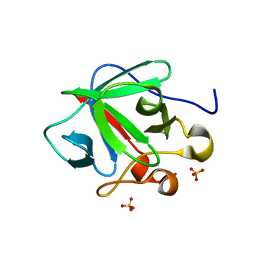 | | Crystal structure of human fibroblast growth factor-1 with mutations Glu81Ser and Lys101Ala | | 分子名称: | Heparin-binding growth factor 1, SULFATE ION | | 著者 | Meher, A.K, Honjo, E, Kuroki, R, Lee, J, Somasundaram, T, Blaber, M. | | 登録日 | 2008-04-07 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Engineering an improved crystal contact across a solvent-mediated interface of human fibroblast growth factor 1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3NAH
 
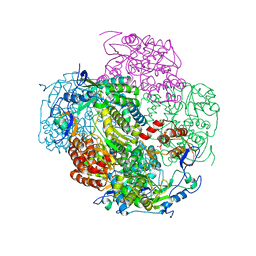 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | 分子名称: | RNA dependent RNA polymerase, SULFATE ION | | 著者 | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | 登録日 | 2010-06-02 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
6K84
 
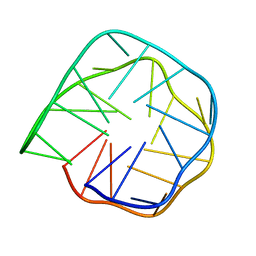 | | Structure of anti-prion RNA aptamer | | 分子名称: | RNA (25-MER) | | 著者 | Mashima, T, Lee, J.H, Hayashi, T, Nagata, T, Kinoshita, M, Katahira, M. | | 登録日 | 2019-06-11 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development and structural determination of an anti-PrPCaptamer that blocks pathological conformational conversion of prion protein.
Sci Rep, 10, 2020
|
|
6KRT
 
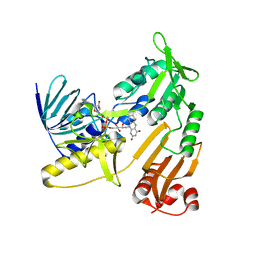 | | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, monodehydroascorbate reductase | | 著者 | Park, A.K, Do, H, Lee, J.H, Kim, H, Choi, W, Kim, I.S, Kim, H.W. | | 登録日 | 2019-08-22 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | monodehydroascorbate reductase, MDHAR, from Antarctic hairgrass Deschampsia antarctica
To Be Published
|
|
3P6I
 
 | |
3P6J
 
 | |
3Q7Y
 
 | |
2HPW
 
 | | Green fluorescent protein from Clytia gregaria | | 分子名称: | Green fluorescent protein | | 著者 | Stepanyuk, G, Liu, Z.J, Vysotski, S.E, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-07-17 | | 公開日 | 2006-09-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of Green Fluorescent Protein from Clytia Gregaria at 1.55 A resolution
To be Published
|
|
3Q7W
 
 | |
3Q7X
 
 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | 著者 | Blaber, M, Lee, J. | | 登録日 | 2011-01-05 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2P2R
 
 | | Crystal structure of the third KH domain of human Poly(C)-Binding Protein-2 in complex with C-rich strand of human telomeric DNA | | 分子名称: | 6-AMINOPYRIMIDIN-2(1H)-ONE, C-rich strand of human telomeric DNA, Poly(rC)-binding protein 2 | | 著者 | James, T.L, Stroud, R.M, Du, Z, Fenn, S, Tjhen, R, Lee, J.K. | | 登録日 | 2007-03-07 | | 公開日 | 2007-06-12 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 A resolution.
Nucleic Acids Res., 35, 2007
|
|
3E2X
 
 | |
3E31
 
 | |
