4HY2
 
 | |
5J4G
 
 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
6LVU
 
 | |
3TD5
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
1X25
 
 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
1F5V
 
 | | STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF A FLAVOPROTEIN FROM ESCHERICHIA COLI THAT REDUCES NITROCOMPOUNDS. ALTERATION OF PYRIDINE NUCLEOTIDE BINDING BY A SINGLE AMINO ACID SUBSTITUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN-INSENSITIVE NADPH NITROREDUCTASE | | Authors: | Kobori, T, Sasaki, H, Lee, W.C, Zenno, S, Saigo, K, Murphy, M.E.P, Tanokura, M. | | Deposit date: | 2000-06-17 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.
J.Biol.Chem., 276, 2001
|
|
1V3Y
 
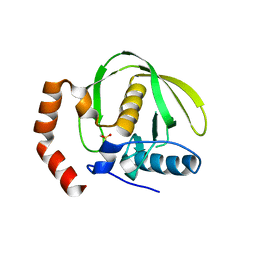 | | The crystal structure of peptide deformylase from Thermus thermophilus HB8 | | Descriptor: | Peptide deformylase | | Authors: | Kamo, M, Kudo, N, Lee, W.C, Ito, K, Motoshim, H, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-07 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The crystal structure of peptide deformylase from Thermus thermophilus HB8
to be published
|
|
2YYS
 
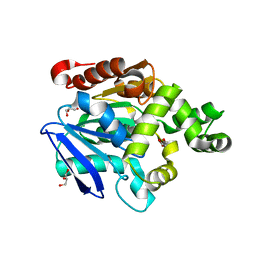 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
3A4I
 
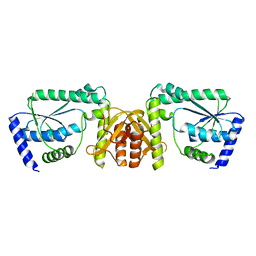 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B | | Authors: | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
5TLC
 
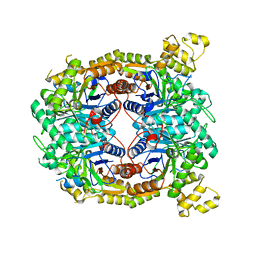 | |
4OH9
 
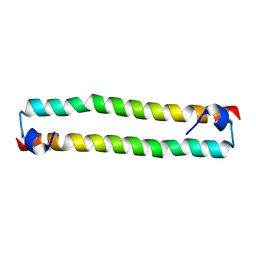 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OH8
 
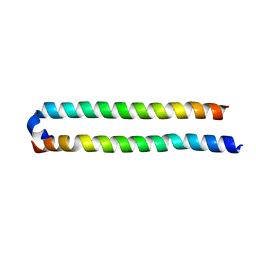 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HE7
 
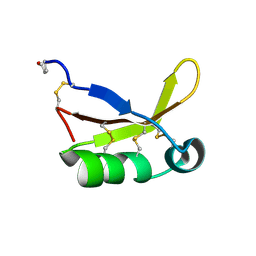 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7V6U
 
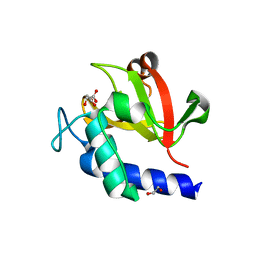 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | The crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6S
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | The crystal structure fre
To be published
|
|
3TRS
 
 | | The crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin-2 heavy chain, Aspergillopepsin-2 light chain, DIMETHYL SULFOXIDE | | Authors: | Sasaki, H, Kubota, K, Lee, W.C, Ohtsuka, J, Kojima, M, Takahashi, K, Tanokura, M. | | Deposit date: | 2011-09-10 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis.
J.Biochem., 152, 2012
|
|
3X0Y
 
 | | Crystal structure of FMN-bound DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC, FLAVIN MONONUCLEOTIDE | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
3X0X
 
 | | Crystal structure of apo-DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
2D7J
 
 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
2D37
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D38
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
