3G0Q
 
 | | Crystal Structure of MutY bound to its inhibitor DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*AP*GP*TP*CP*TP*T)-3', A/G-specific adenine glycosylase, ... | | Authors: | Lee, S, Verdine, G.L. | | Deposit date: | 2009-01-28 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SVP
 
 | | SINDBIS VIRUS CAPSID PROTEIN | | Descriptor: | SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Rossmann, M.G. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly.
Structure, 4, 1996
|
|
5HHI
 
 | | Structure of human DNA polymerase beta Host-Guest complexed with CBZ-platinated N7-G | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Lee, S, Koag, M.-C. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.517 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
5VAN
 
 | | Crystal Structure of Beta-Klotho | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAK
 
 | | Crystal Structure of Beta-Klotho, Domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5IP5
 
 | |
5VAQ
 
 | | Crystal Structure of Beta-Klotho in Complex with FGF21CT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, Fibroblast growth factor 21, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5CUS
 
 | | Crystal Structure of sErbB3-Fab3379 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab LC region of KTN3379, IgG H chain, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2015-07-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4P2H
 
 | |
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
3PMR
 
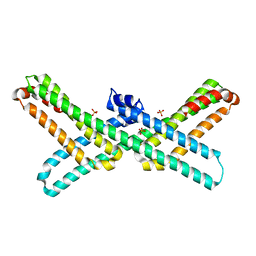 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
1UZK
 
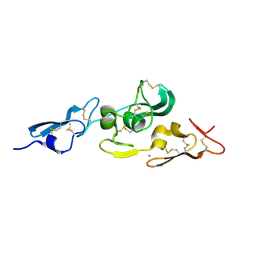 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Ca bound to cbEGF23 domain only | | Descriptor: | CALCIUM ION, FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-13 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
1UZP
 
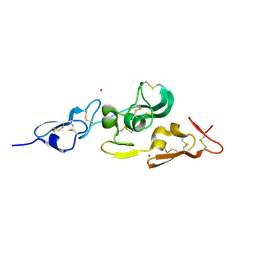 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Sm bound form cbEGF23 domain only. | | Descriptor: | FIBRILLIN-1, SAMARIUM (III) ION | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-15 | | Release date: | 2004-04-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
1UZJ
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, holo form. | | Descriptor: | CALCIUM ION, FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
1UZQ
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, apo form cbEGF23 domain only. | | Descriptor: | FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-15 | | Release date: | 2004-04-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
7YMO
 
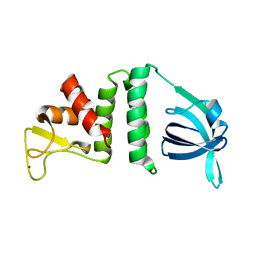 | |
5XD9
 
 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
1P7G
 
 | | Crystal structure of superoxide dismutase from Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Superoxide dismutase | | Authors: | Lee, S, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2003-05-01 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of superoxide dismutase from Pyrobaculum aerophilum presents a challenging case in molecular replacement with multiple molecules, pseudo-symmetry and twinning.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2KMK
 
 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | Authors: | Lee, S, Wu, Z. | | Deposit date: | 2009-07-30 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
2FID
 
 | |
2FIF
 
 | |
