7CT7
 
 | | FECH - inhibitor complex 2 | | Descriptor: | 2-[[(4-chlorophenyl)amino]methyl]-5-propyl-6H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Lee, S.J, Park, J. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | FECH - inhibitor complex 1
to be published
|
|
4R6W
 
 | |
3UJ6
 
 | | SeMet Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAM and PO4 | | Descriptor: | PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
5IP5
 
 | |
3UJB
 
 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAH and phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphoethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
2FID
 
 | |
2FIF
 
 | |
4GAM
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit | | Descriptor: | FE (III) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Lee, S.J, Lippard, S.J, Cho, U.-S. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Control of substrate access to the active site in methane monooxygenase.
Nature, 494, 2013
|
|
3UJ7
 
 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAM and PO4 | | Descriptor: | PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
2KMK
 
 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | Authors: | Lee, S, Wu, Z. | | Deposit date: | 2009-07-30 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
7KRG
 
 | |
2OJM
 
 | |
2OJN
 
 | |
2OJO
 
 | |
7KQV
 
 | |
6OIT
 
 | | CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1) | | Descriptor: | Protein CHROMATIN REMODELING 35, Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6OIS
 
 | | CryoEM structure of Arabidopsis DR complex (DMS3-RDM1) | | Descriptor: | Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
2QHU
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTANAL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2ARU
 
 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
7LLQ
 
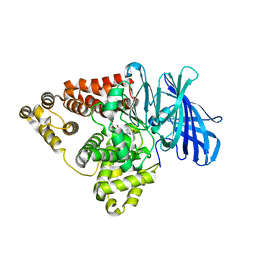 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
4W5P
 
 | | Prp peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4W71
 
 | | Crystal structure of a prion peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-08-21 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
