5ACO
 
 | | Cryo-EM structure of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 ENVELOPE GLYCOPROTEIN, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy.
Structure, 23, 2015
|
|
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
4QXL
 
 | | Crystal Structure of FLHE | | Descriptor: | Flagellar protein flhE | | Authors: | Lee, J, Monzingo, A.F, Keatinge-Clay, A.T, Harshey, R.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-01-14 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Structure of Salmonella FlhE, Conserved Member of a Flagellar Type III Secretion Operon.
J.Mol.Biol., 427, 2015
|
|
3HGT
 
 | |
3HGQ
 
 | |
4MRN
 
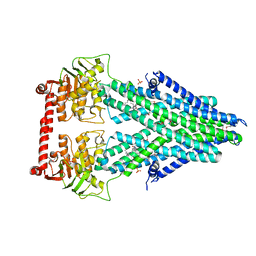 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRS
 
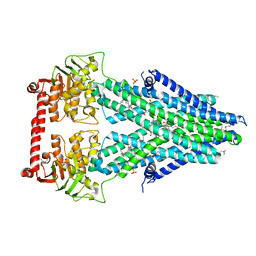 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRR
 
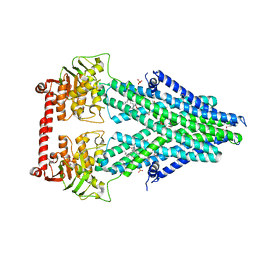 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRV
 
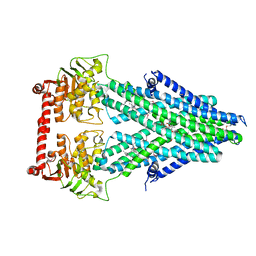 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MVC
 
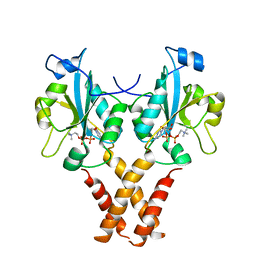 | | Crystal Structure of a Mammalian Cytidylyltransferase | | Descriptor: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Lee, J, Cornell, R.B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
5XWB
 
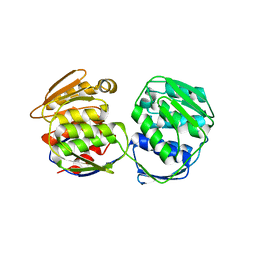 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
1YTO
 
 | | Crystal Structure of Gly19 deletion Mutant of Human Acidic Fibroblast Growth Factor | | Descriptor: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-02-10 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
1Z4S
 
 | |
3H2V
 
 | | Human raver1 RRM1 domain in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
3O3Q
 
 | |
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
3EQA
 
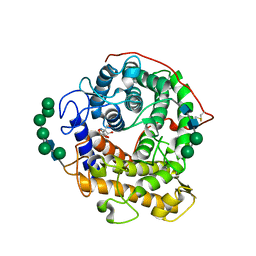 | | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glucoamylase, ... | | Authors: | Lee, J, Paetzel, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the catalytic domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1MZM
 
 | |
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3O4A
 
 | |
3PD7
 
 | |
4Q16
 
 | |
7Y7O
 
 | |
6BQ1
 
 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
